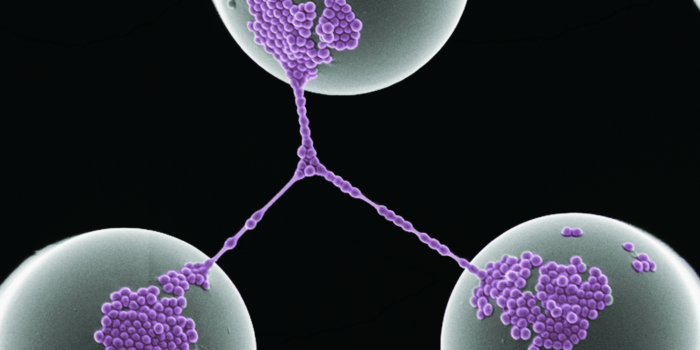
 Cell Adhesion and Network
Cell Adhesion and Network Application of NLP and Deep Learning in Bioinformatics
Application of NLP and Deep Learning in Bioinformatics Deep Learning in Biology and Medicine
Deep Learning in Biology and Medicine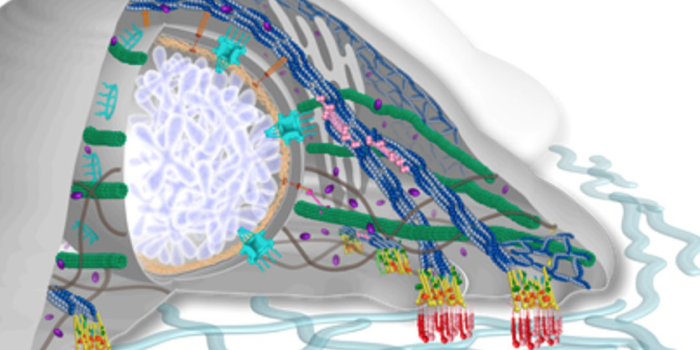 Looking “Under the Hood” of Cellular Mechanotransduction
Looking “Under the Hood” of Cellular Mechanotransduction Deep Proteomics: Protein Vectors (ProtVec and ProtVecX) and DiMotif
Deep Proteomics: Protein Vectors (ProtVec and ProtVecX) and DiMotif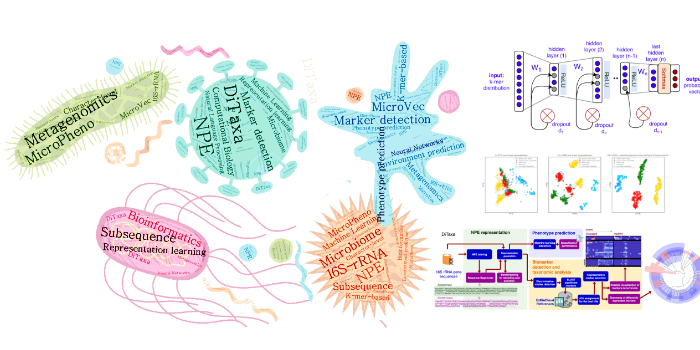 Microbial Informatics with MicroPheno and DiTaxa
Microbial Informatics with MicroPheno and DiTaxa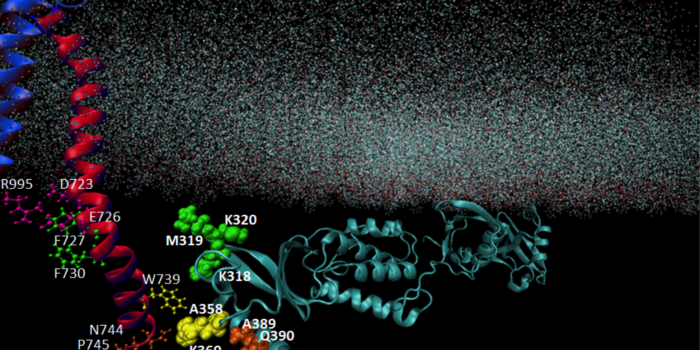 Cell Adhesion: Inside-Out Activation of Integrin via Talin
Cell Adhesion: Inside-Out Activation of Integrin via Talin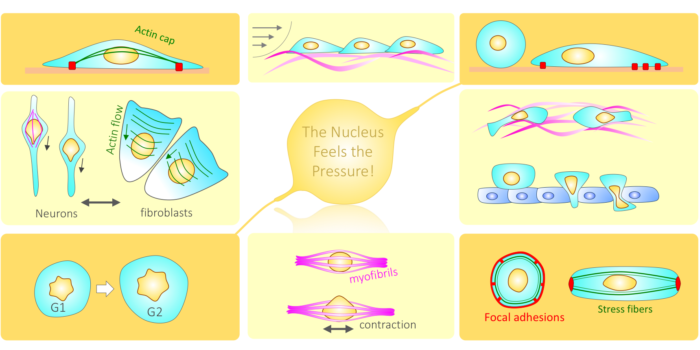 The nucleus feels the force!
The nucleus feels the force!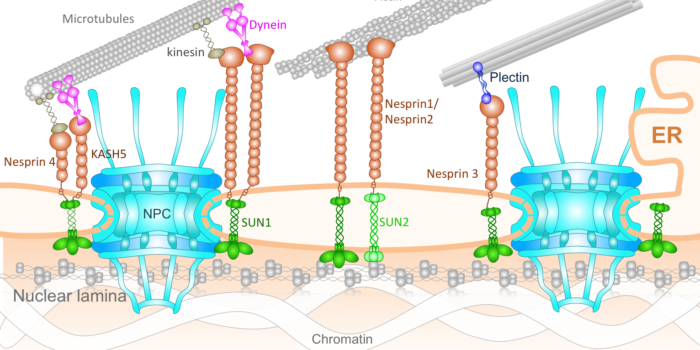 Nucleo-Cytoskeletal Connection
Nucleo-Cytoskeletal Connection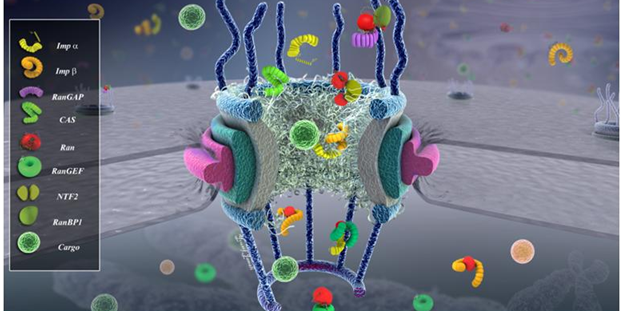 Nuclear Pore Biomechanics and Mechanobiology of Nucleocytoplasmic Transport
Nuclear Pore Biomechanics and Mechanobiology of Nucleocytoplasmic Transport Free Energy Calculations Shed Light on the Nuclear Pore Complex’s Selective Barrier Nature.
Free Energy Calculations Shed Light on the Nuclear Pore Complex’s Selective Barrier Nature.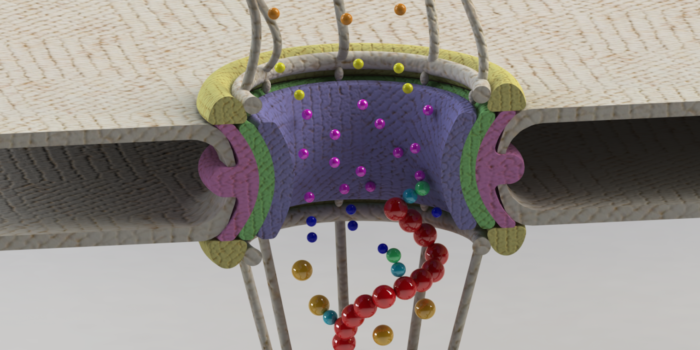 mRNA export out of the nuclear pore complex
mRNA export out of the nuclear pore complex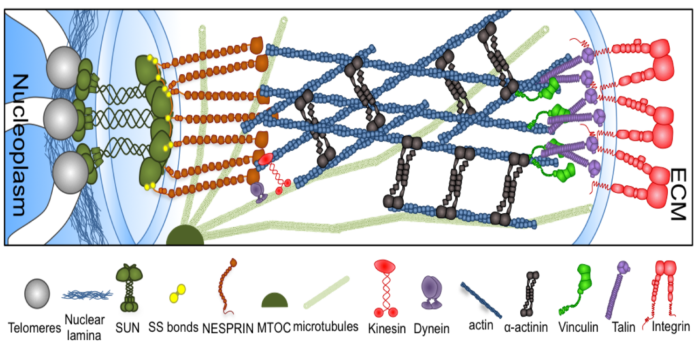 Mechanotransduction Pathways, from ECM to Nucleus
Mechanotransduction Pathways, from ECM to Nucleus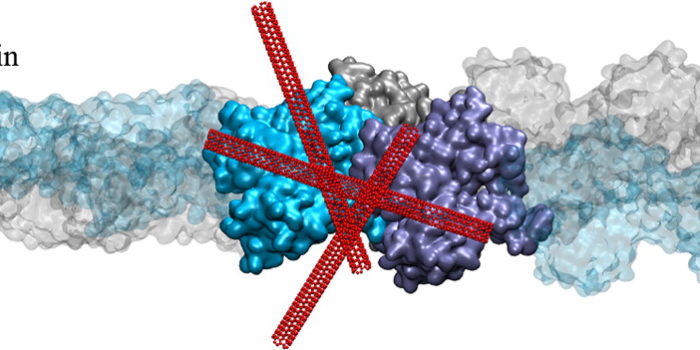 Actin Reorganization through Dynamic Interactions with Single-Wall Carbon Nanotubes (CNTs)
Actin Reorganization through Dynamic Interactions with Single-Wall Carbon Nanotubes (CNTs)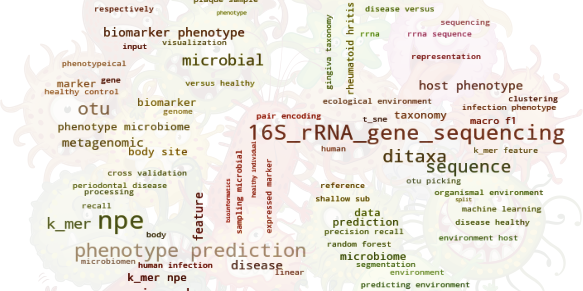 Life Language Processing for Deep Metagenomics
Life Language Processing for Deep Metagenomics Life Language Processing for Deep Proteomics
Life Language Processing for Deep Proteomics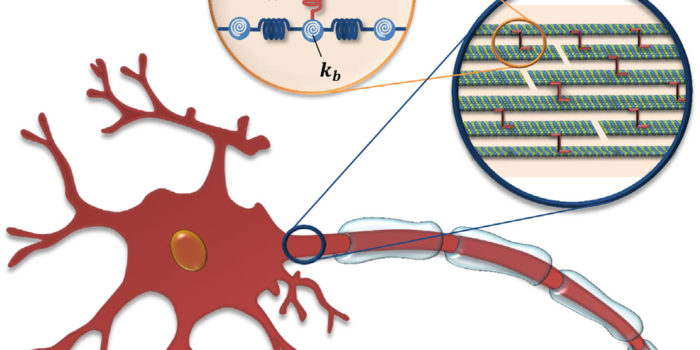 Neuronal Axon Biomechanics
Neuronal Axon Biomechanics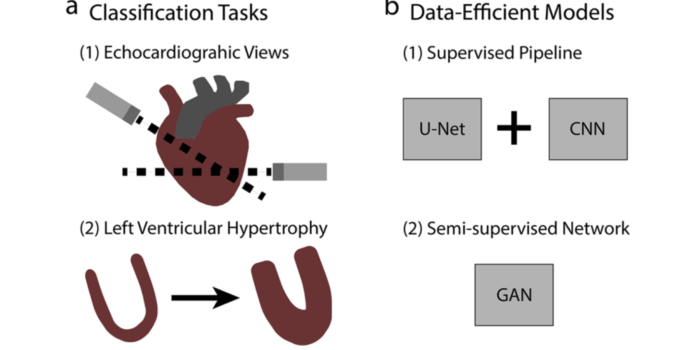 Deep echocardiography: Deep learning towards automated diagnosis of cardiac disease
Deep echocardiography: Deep learning towards automated diagnosis of cardiac disease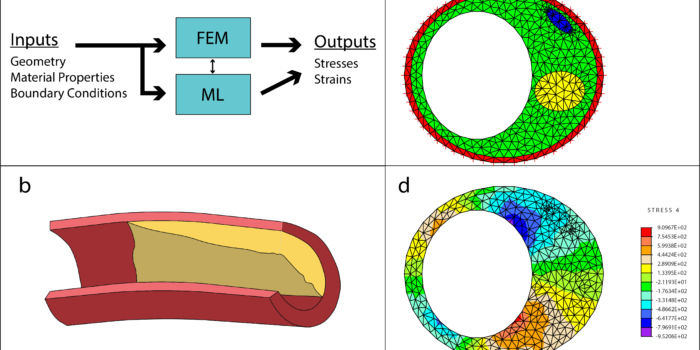 Bridging finite element and machine learning modeling: stress prediction of arterial walls in atherosclerosis
Bridging finite element and machine learning modeling: stress prediction of arterial walls in atherosclerosis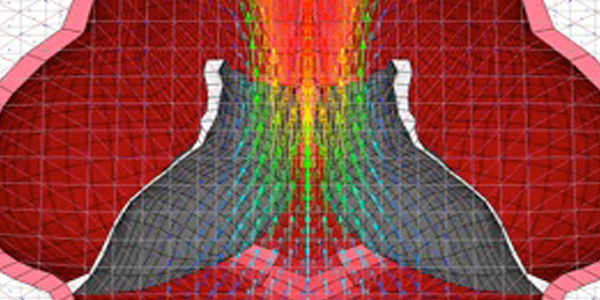 Biomechanics of Heart Valves
Biomechanics of Heart Valves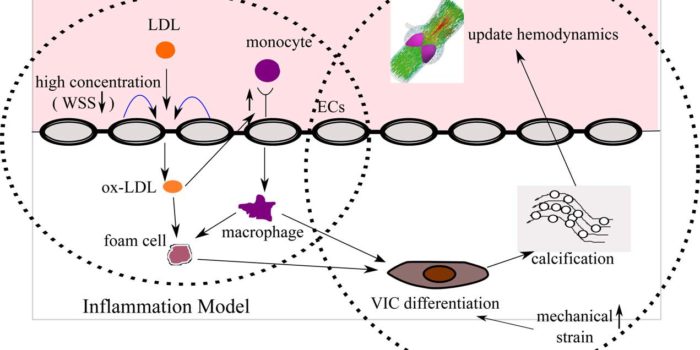 Aortic Valve Calcification
Aortic Valve Calcification
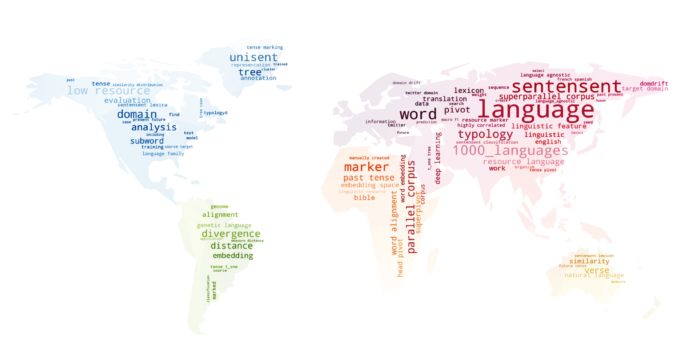 Multilingual Language Processing
Multilingual Language Processing
 Thank you for supporting our research!
Thank you for supporting our research!
Welcome to Mofrad Laboratory! Our mission is to understand the molecular basis of human diseases via state-of-the-art molecular biophysics and computational biology approaches.