Publications
Books
Journal Papers
 |
Deciphering the intrinsically disordered characteristics of the FG-Nups through the lens of polymer physics Matsuda A, Mansour A, Mofrad MRK. Molecular Biology of the Cell, Nucleus. (KEYWORDS: Nuclear Pore Complex, Nucleocytoplasmic Transport) |
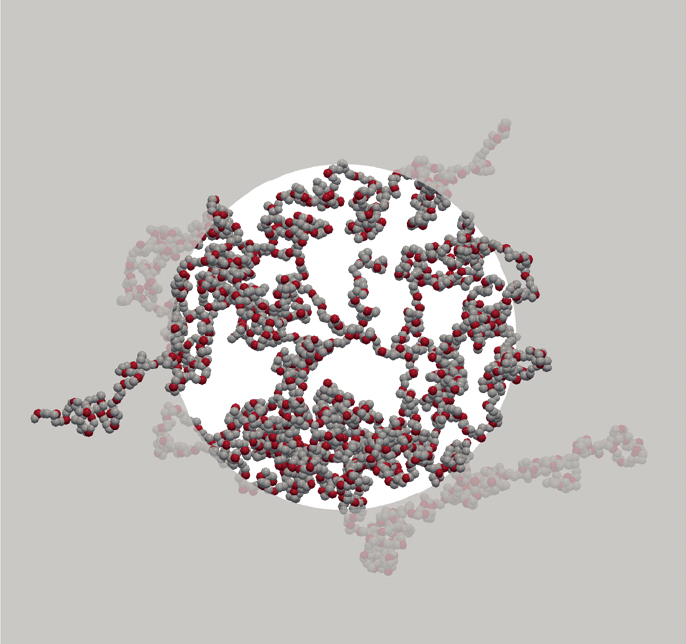 |
Regulating transport efficiency through the nuclear pore complex: the role of binding affinity with FG-Nups Matsuda A, Mofrad MRK Molecular Biology of the Cell, In Press. (KEYWORDS: Nuclear Pore Complex, Nucleocytoplasmic Transport) |
 |
Nanoscale In silico and In vitro Modeling of Lipid Bilayers for Curvature Induction and Sensing Ghafar Yerima, Ching-Ting Tsai, Chih-hao Lu, Bianxiao Cui*, Zeinab Jahed*, Mohammad R.K. Mofrad*. npj Biological Physics & Mechanics, In Press. (KEYWORDS: BAR domain proteins, Cell Membrane Curvature, Focal Adhesions) |
 |
Bacterial community dynamics as a result of growth-yield trade-off and multispecies metabolic interactions toward understanding the gut biofilm niche Amin Valiei, Andrew Dickson, Javad Aminian-Dehkordi & Mohammad R. K. Mofrad BMC Microbiology, In Press. (KEYWORDS: Microbiomemes, Bacterial Communities) |
 |
Metabolic interactions shape emergent biofilm structures in a conceptual model of gut mucosal bacterial communities Amin Valiei, Andrew Dickson, Javad Aminian-Dehkordi & Mohammad R. K. Mofrad npj Biofilms and Microbiomes volume 10, Article number: 99 (2024) ; doi: 10.1038/s41522-024-00572-y (KEYWORDS: Microbiomemes, Bacterial Communities) |
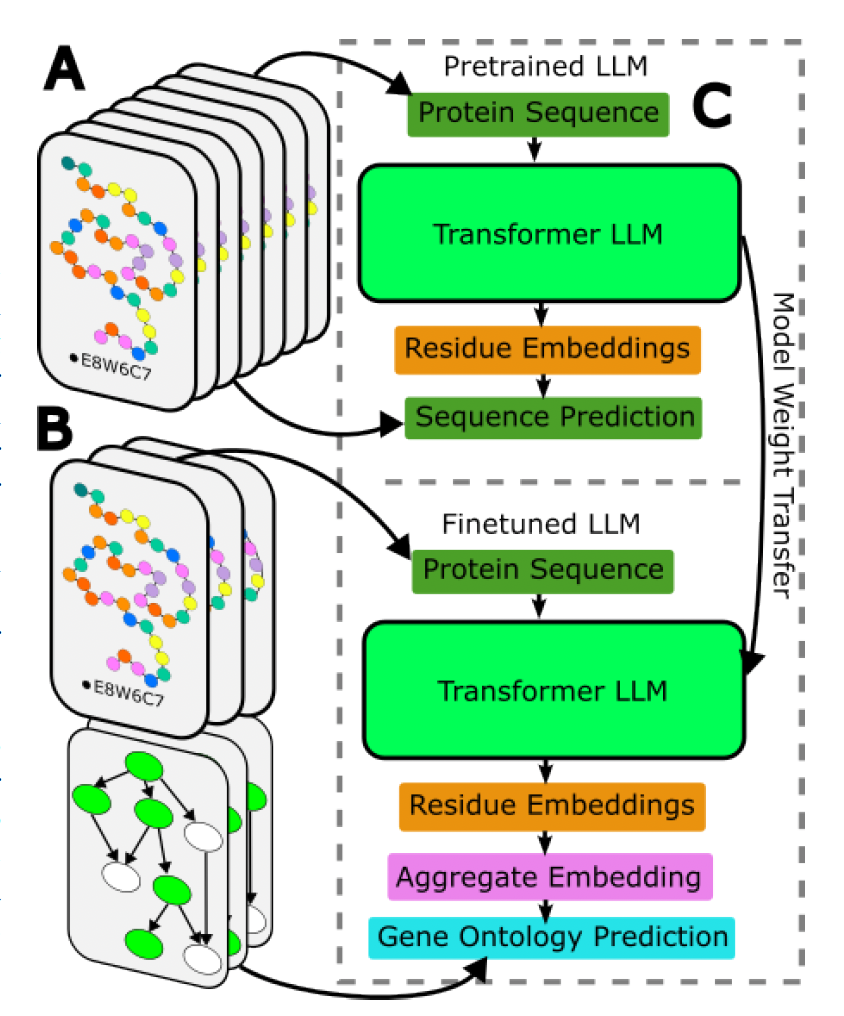 |
Fine-tuning Protein Embeddings for Functional Similarity Evaluations AM Dickson, MRK Mofrad. Bioinformatics, In Press. |
 |
Integrin Mechanosensing relies on Pivot-clip Mechanism to Reinforce Cell Adhesion AR Montes, A Barroso, W Wang, GD O’Connell, AB Tepole, MRK Mofrad. Biophysical Journal, In Press. |
 |
HGTDR: Advancing Drug Repurposing with Heterogeneous Graph Transformers A Gharizadeh, K Abbasi, A Ghareyazi, MRK Mofrad, HR Rabiee. Bioinformatics, In Press. |
 |
Biomechanics and Mechanobiology: Mechano-Genomics D Leckband, MRK Mofrad Current Opinion in Biomedical Engineering, 100536, 2024. |
 |
Deep Learning for Perfusion Cerebral Blood Flow (CBF) and Volume (CBV) Predictions and Diagnostics S Talebi, S Gai, A Sossin, V Zhu, E Tong, MRK Mofrad Annals of Biomedical Engineering, 1-8, 2024. |
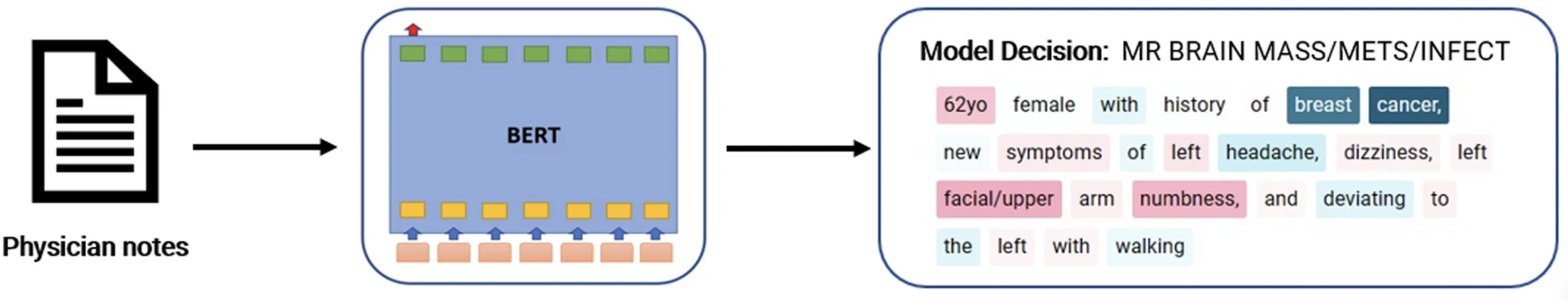 |
Exploring the performance and explainability of fine-tuned BERT models for neuroradiology protocol assignment S Talebi, E Tong, A Li, G Yamin, G Zaharchuk, MRK Mofrad BMC Medical Informatics and Decision Making 24 (1), 40, 2024, 2024. |
 |
Antimicrobial peptide interactions with bacterial cell membranes M Khavani, A Mehranfar, MRK Mofrad Journal of Biomolecular Structure and Dynamics, 1-14, 2024. |
 |
Enhanced HP1α homodimer interaction via force-induced salt bridge formation: implications for chromatin crosslinking and phase separation S Tsukamoto, M Khavani, N Domkam, MRK Mofrad RSC Mechanochemistry, 2024. Featured in RSC Mechanochemistry Showcase as one of the top Editor’s Choice articles. |
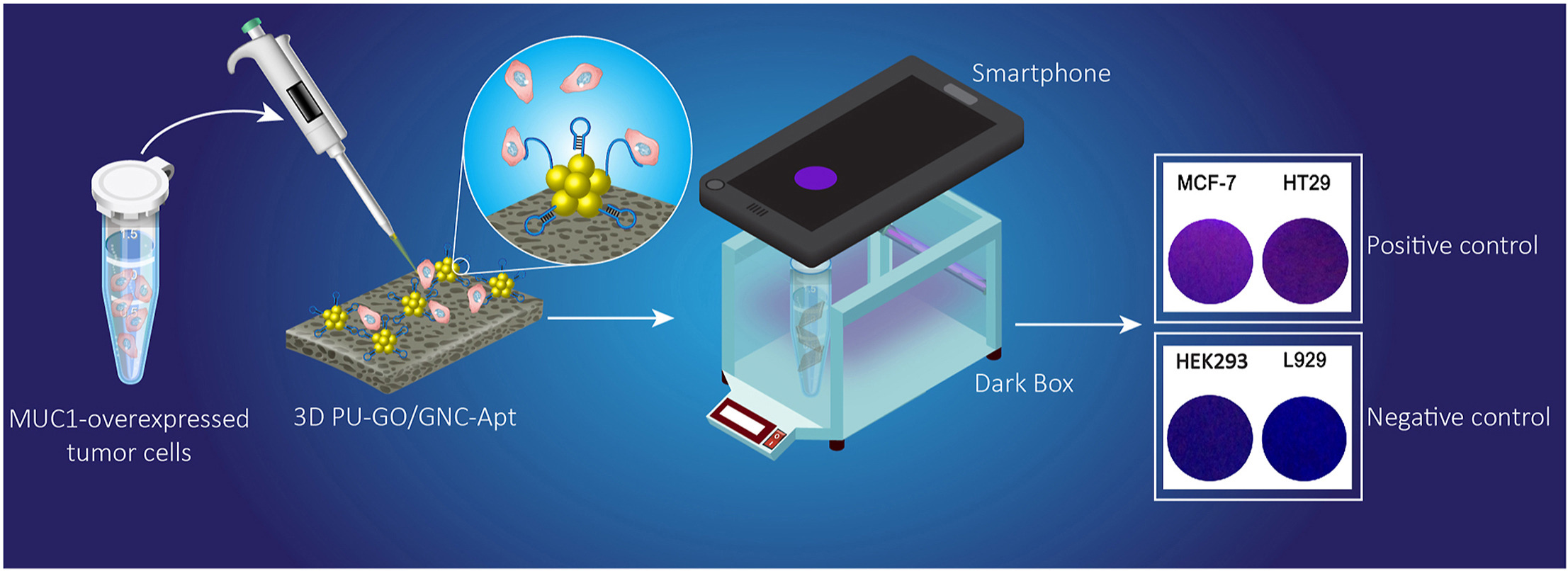 |
Smartphone-assisted lab-in-a-tube device using gold nanocluster-based aptasensor for detection of MUC1-overexpressed tumor cells A Sanati, Y Esmaeili, M Khavani, E Bidram, A Rahimi, A Dabiri, … Analytica Chimica Acta 1252, 341017, 2023. |
 |
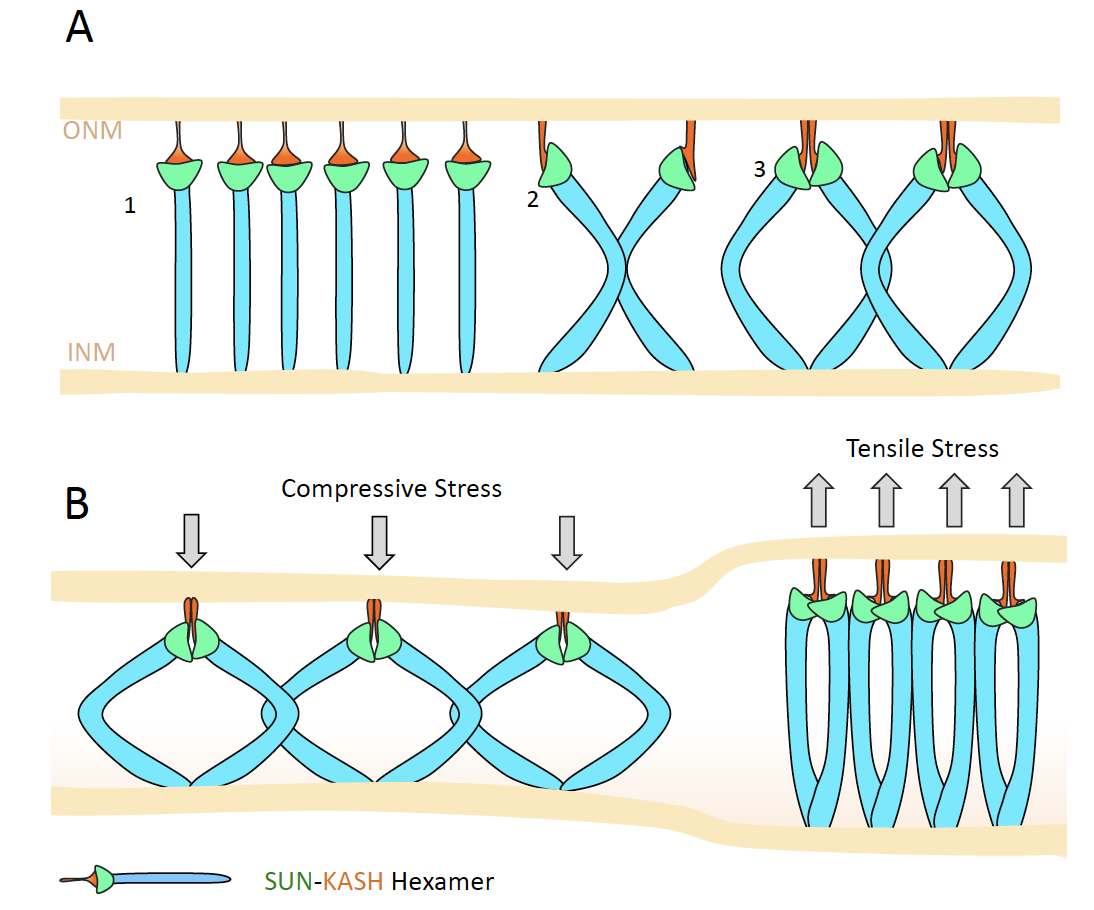
Force transmission and SUN-KASH higher-order assembly in the LINC complex models G Yerima, N Domkam, J Ornowski, Z Jahed, MRK Mofrad Biophysical Journal 122 (23), 4582-4597, 2023. |
 |
Multiscale computational framework to investigate integrin mechanosensing and cell adhesion AR Montes, G Gutierrez, A Buganza Tepole, MRK Mofrad Journal of Applied Physics 134 (11), 2023. |
 |
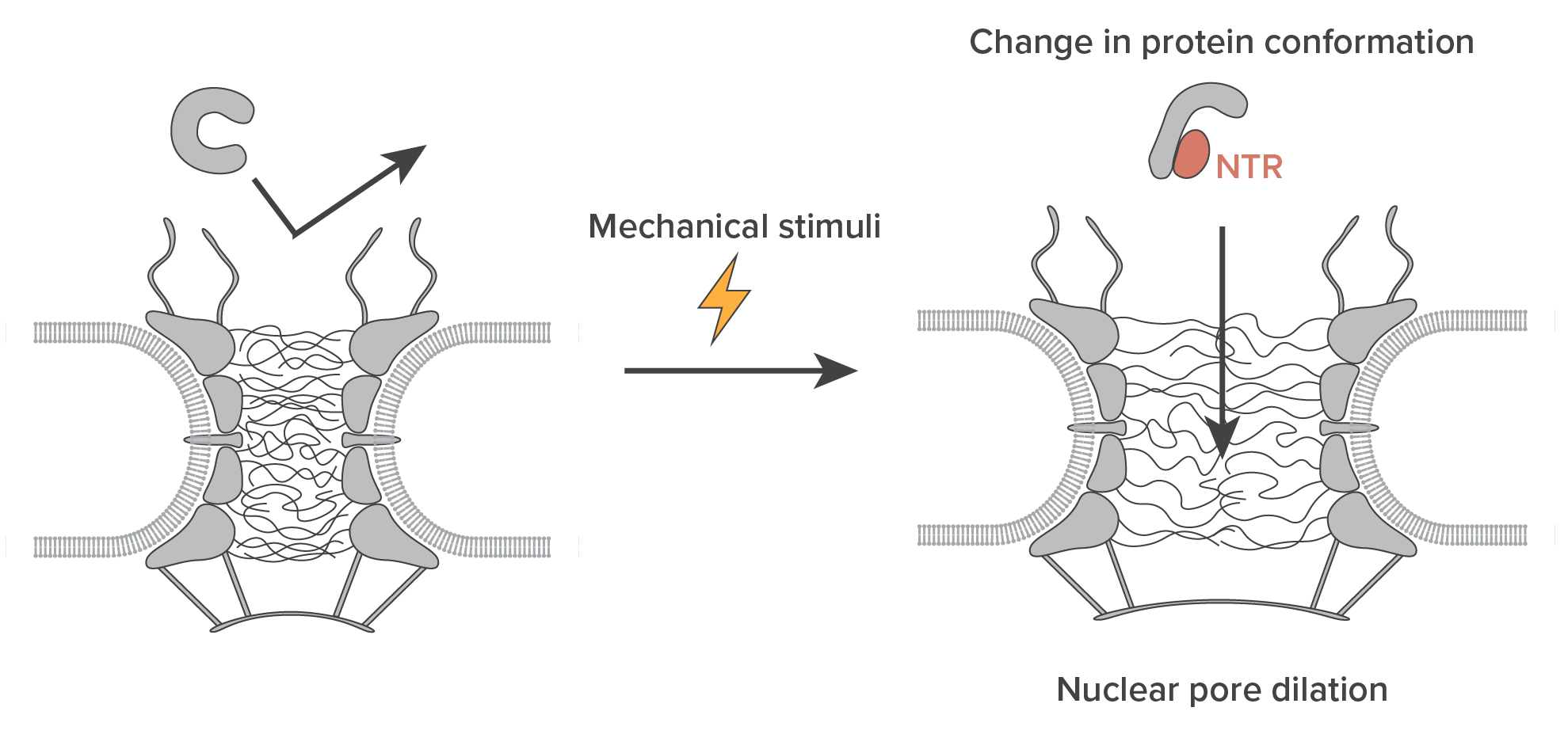
Role of Pore Dilation in Molecular Transport through the Nuclear Pore Complex: Insights from Polymer Scaling Theory A Matsuda, MRK Mofrad arXiv preprint arXiv:2307.07959, 2023. |
 |
Beyond the Hype: Assessing the Performance, Trustworthiness, and Clinical Suitability of GPT3. 5 S Talebi, E Tong, MRK Mofrad arXiv preprint arXiv:2306.15887, 2023. |
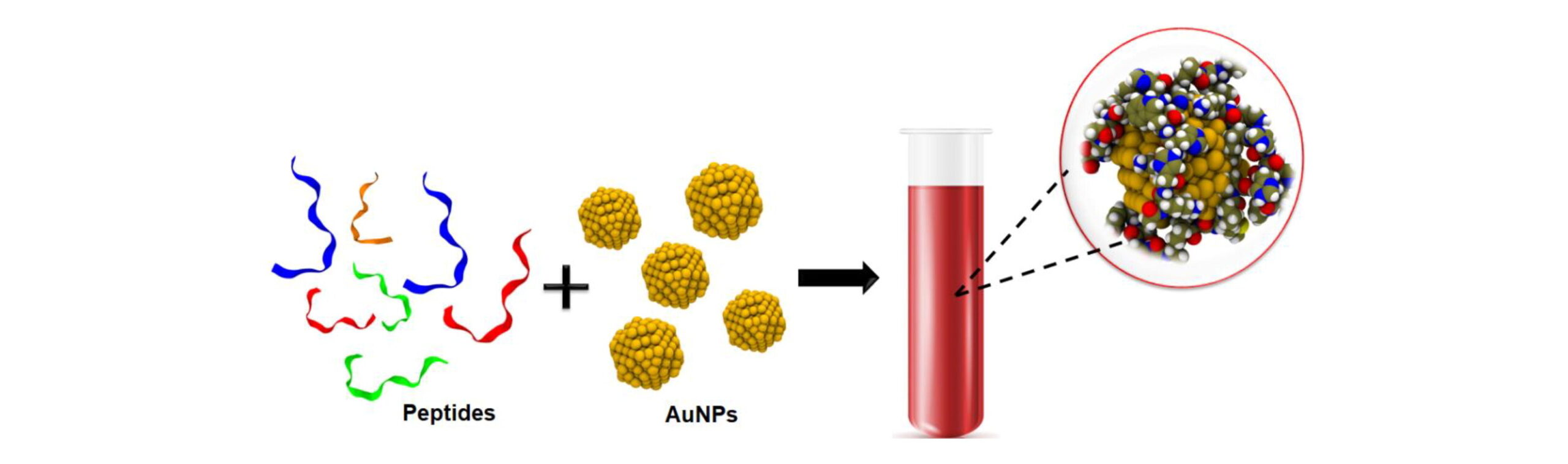 |
On the interactions of peptides with gold nanoparticles: effects of sequence and size M Khavani, A Mehranfar, MRK Mofrad. Journal of Biomolecular Structure and Dynamics, 1-13, 2023 |
 |
Adsorption Process of Various Antimicrobial Peptides on Different Surfaces of Cellulose A Mehranfar, M Khavani, MRK Mofrad ACS Applied Bio Materials 6 (3), 1041-1053, 2023. |
 |
Gut-on-a-chip models for dissecting the gut microbiology and physiology A Valiei, J Aminian-Dehkordi, MRK Mofrad APL bioengineering 7 (1), 2023. |
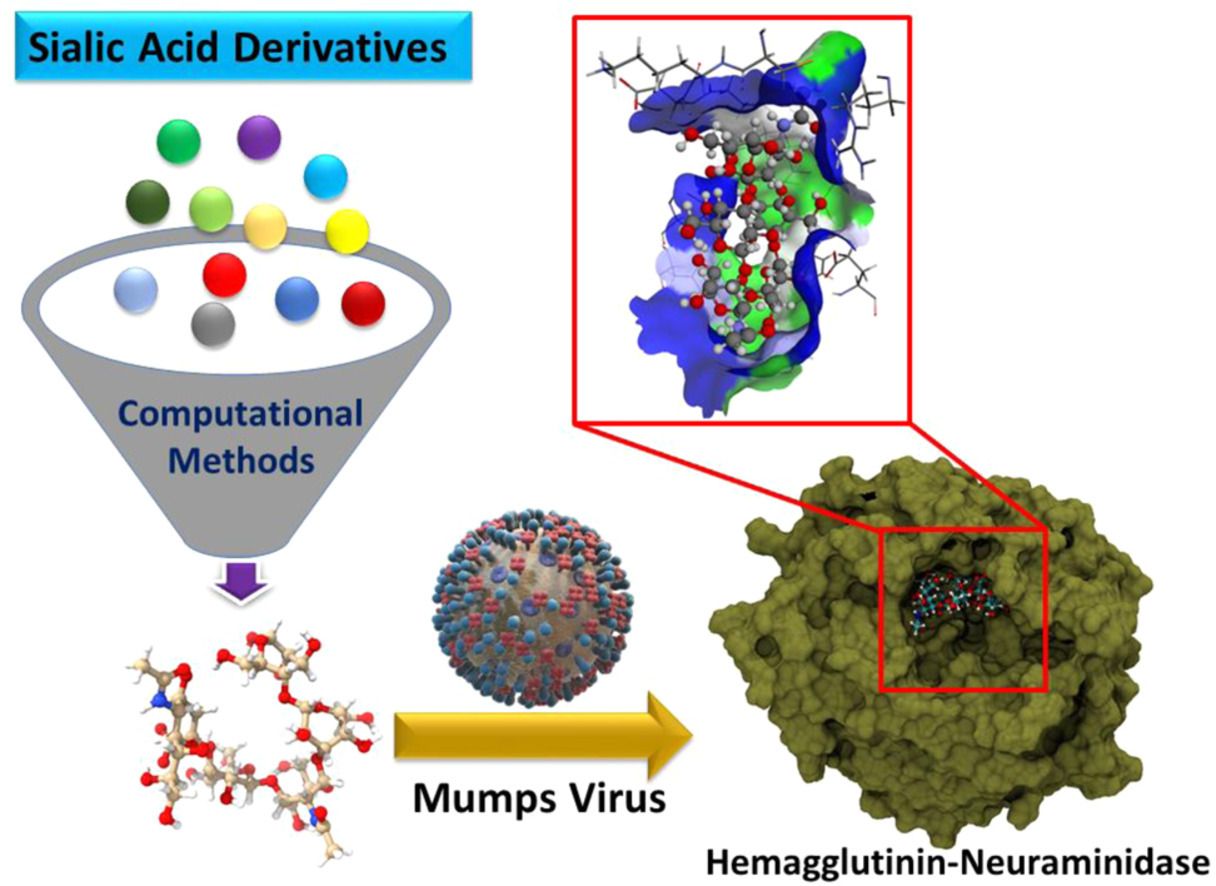 |
On the potentials of sialic acid derivatives as inhibitors for the mumps virus: A molecular dynamics and quantum chemistry investigation M Khavani, A Mehranfar, MRK Mofrad Virus Research 326, 199050, 2023. |
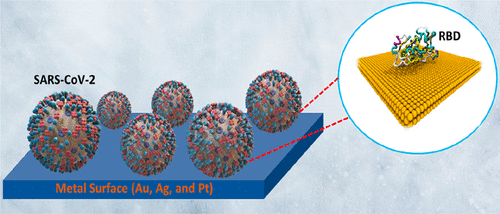 |
On the Sensitivity and Affinity of Gold, Silver, and Platinum Surfaces against the SARS-CoV-2 Virus: A Comparative Computational Study M Khavani, A Mehranfar, MRK Mofrad Journal of Chemical Information and Modeling 63 (4), 1276-1292, 2023. |
 |
GO Bench: shared hub for universal benchmarking of machine learning-based protein functional annotations A Dickson, E Asgari, AC McHardy, MRK Mofrad Bioinformatics 39 (2), btad081, 2023. |
 |
Mercapto-pyrimidines are reversible covalent inhibitors of the papain-like protease (PLpro) and inhibit SARS-CoV-2 (SCoV-2) replication T Bajaj, E Wehri, RK Suryawanshi, E King, KS Pardeshi, K Behrouzi, … RSC advances 13 (26), 17667-17677, 2023. |
 |
A covalent inhibitor targeting the papain-like protease from SARS-CoV-2 inhibits viral replication H Han, AV Gracia, JJ Røise, L Boike, K Leon, U Schulze-Gahmen, … RSC advances 13 (16), 10636-10641, 2023. |
 |
Effects of ionic liquids on the stabilization process of gold nanoparticles M Khavani, A Mehranfar, MRK Mofrad The Journal of Physical Chemistry B 126 (46), 9617-9631, 2022. |
 |
Emerging computational paradigms to address the complex role of gut microbial metabolism in cardiovascular diseases J Aminian-Dehkordi, A Valiei, MRK Mofrad Frontiers in cardiovascular medicine 9, 987104, 2022. |
 |
Design of a flexing organ-chip to model in situ loading of the intervertebral disc JP McKinley, AR Montes, MN Wang, AR Kamath, G Jimenez, J Lim, … Biomicrofluidics 16 (5), 2022. |
 |
On modeling the multiscale mechanobiology of soft tissues: Challenges and progress Y Guo, MRK Mofrad, AB Tepole Biophysics Reviews 3 (3), 2022. |
 |
Decoding the effect of hydrostatic pressure on TRPV1 lower-gate conformation by molecular-dynamics simulation MHB Zamri, Y Ujihara, M Nakamura, MRK Mofrad, S Sugita International journal of molecular sciences 23 (13), 7366, 2022. |
 |
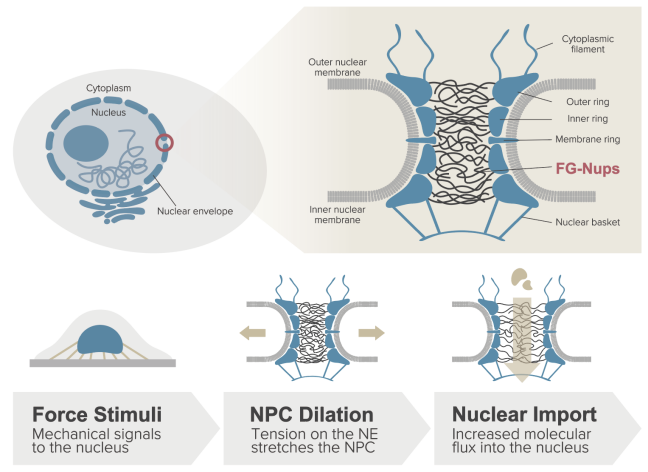
On the Nuclear Pore Complex and Its Emerging Role in Cellular Mechanotransduction Atsushi Matsuda, Mohammad R. K. Mofrad. APL Bioengineering (special issue on Mechanobiology of the Cell Nucleus), 6, 011504 (2022); doi: 10.1063/5.0080480 (KEYWORDS: Nuclear Pore Complex, Mechanotransduction) |
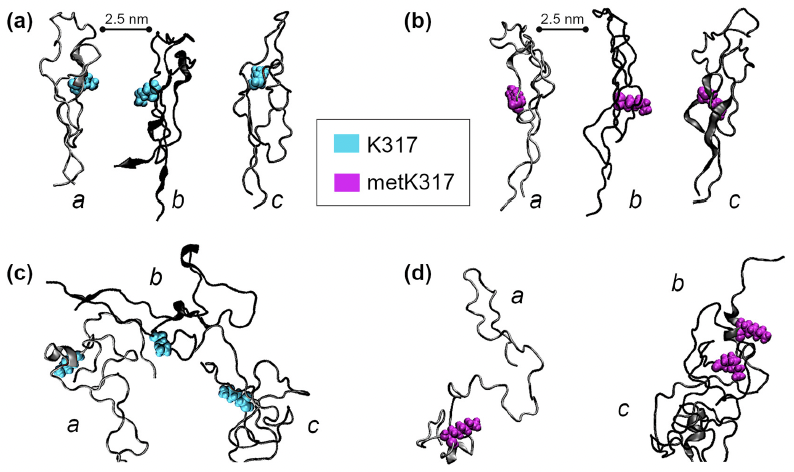 |
Methylation at a conserved lysine residue modulates tau assembly and cellular functions Hengameh Shams, Atsuko Matsunaga, Qin Ma, Mohammad R.K. Mofrad, and Alessandro Didonna Molecular and Cellular Neuroscience 120 (2022) 103707. (KEYWORDS: Tau Protein Methylation, Molecular Dynamics) |
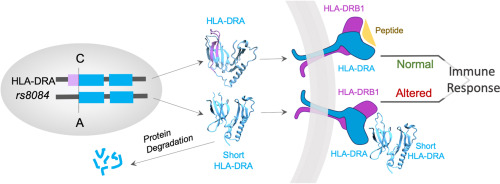 |
A short HLA-DRA isoform binds the HLA-DR2 heterodimer on the outer domain of the peptide-binding site Hengameh Shams, Jill A. Hollenbach, Atsuko Matsunaga, Mohammad R.K. Mofrad, Jorge R. Oksenberg a, and Alessandro Didonna Archives of Biochemistry and Biophysics 719 (2022) 109156 (KEYWORDS: immune system, human leukocyte antigen, antigen-presenting cells, Molecular Dynamics) |
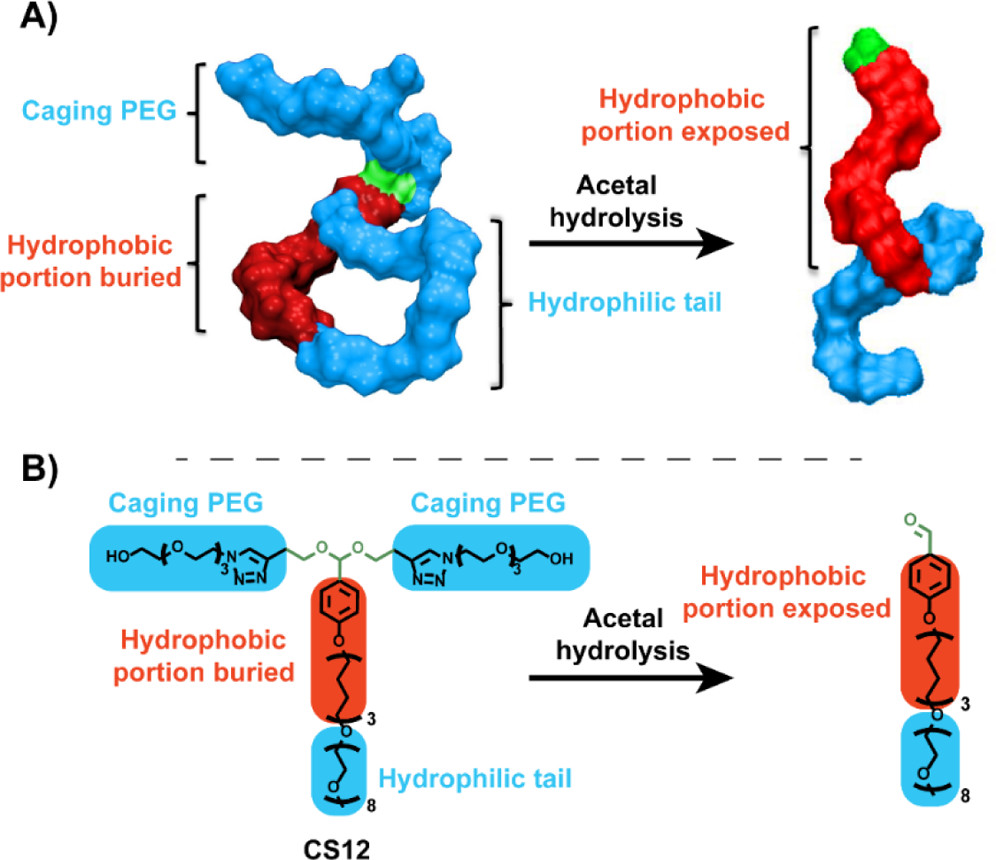 |
Acid-Sensitive Surfactants Enhance the Delivery of Nucleic Acids Joachim Justad Røise, Hesong Han, Jie Li, D. Lucas Kerr, Chung Taing, Kamyar Behrouzi, Maomao He, Emily Ruan, Lienna Y. Chan, Eli M. Espinoza, Sören Reinhard, Kanav Thakker, Justin Kwon, Mohammad R. K. Mofrad, and Niren Murthy. Molecular Pharmaceutics. 19(1):67–79, 20222 (KEYWORDS: Nucleic Acids) |
 |
Characterizing Binding Interactions That Are Essential for Selective Transport through the Nuclear Pore Complex Lennon KM, Soheilypour M, Peyro M, Wakefield DL, Choo GE, Mofrad MRK* and Jovanovic-Talisman T*. International Journal of Molecular Sciences. 22(19):10898, 2021 (KEYWORDS: Nuclear pore complex, Nucleoporins) |
 |
Free Energy Calculations Shed Light on the Nuclear Pore Complex’s Selective Barrier Nature. A. Matsuda, MRK Mofrad. Biophysical Journal, 20(17):3628-3640, SEPTEMBER 07, 2021 [Cover] (KEYWORDS: Nuclear pore complex, free energy, Selective Permeable.Entry in the Biophysical Journal Blog: Nature’s Most Exquisite Nanopore and Its Vital Role in Cell Biology |
 |
Nucleoporins’ exclusive amino acid sequence features regulate their transient interaction with and selectivity of cargo complexes in the nuclear pore. Peyro M, Dickson AM, MRK Mofrad. Molecular Biology of the Cell, 2021, In Press. (KEYWORDS: Nuclear pore complex, Selective Permeable.) |
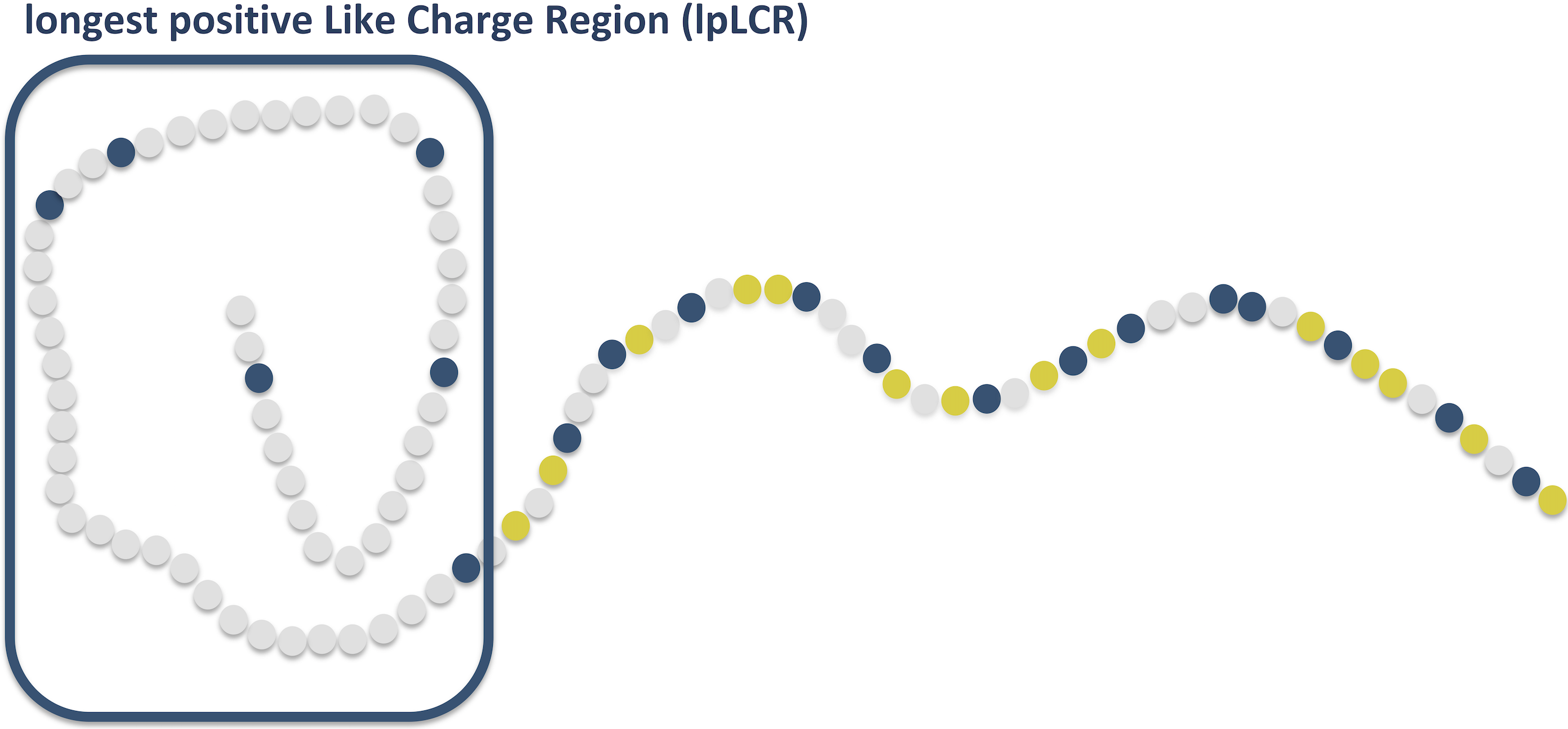 |
FG Nucleoporins feature unique amino acid sequence patterns that distinguish them from other IDPs. M Peyro*, M Soheilypour*, VS Nibber, AM Dickson, MRK Mofrad. Biophysical Journal, 120(16):3382-3391, AUGUST 17, 2021 (KEYWORDS: Nuclear pore complex, intrinsically disordered proteins.) |
 |
TripletProt: Deep Representation Learning of Proteins based on Siamese Networks Nourani E, Asgari E, McHardy AC, and Mofrad MRK. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2021 (In Press). (KEYWORDS: Proteomics, Deep Learning, Neural Networks.) |
 |
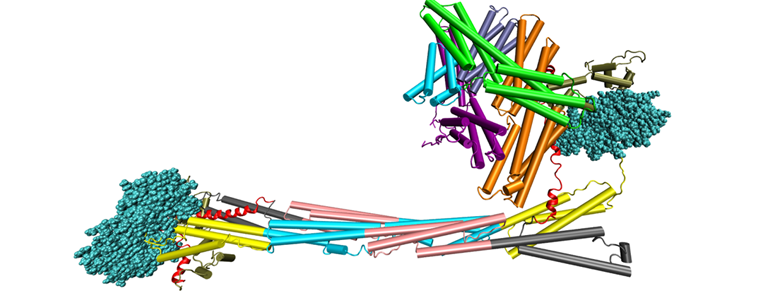
Molecular models of LINC complex assembly at the nuclear envelope. Z Jahed*, N Domkam, J Ornowski, G Yerima, MRK Mofrad*. Journal of Cell Science 134 (12), jcs258194 (KEYWORDS: LINC Complex, Nuclear Mechanotransduction) |
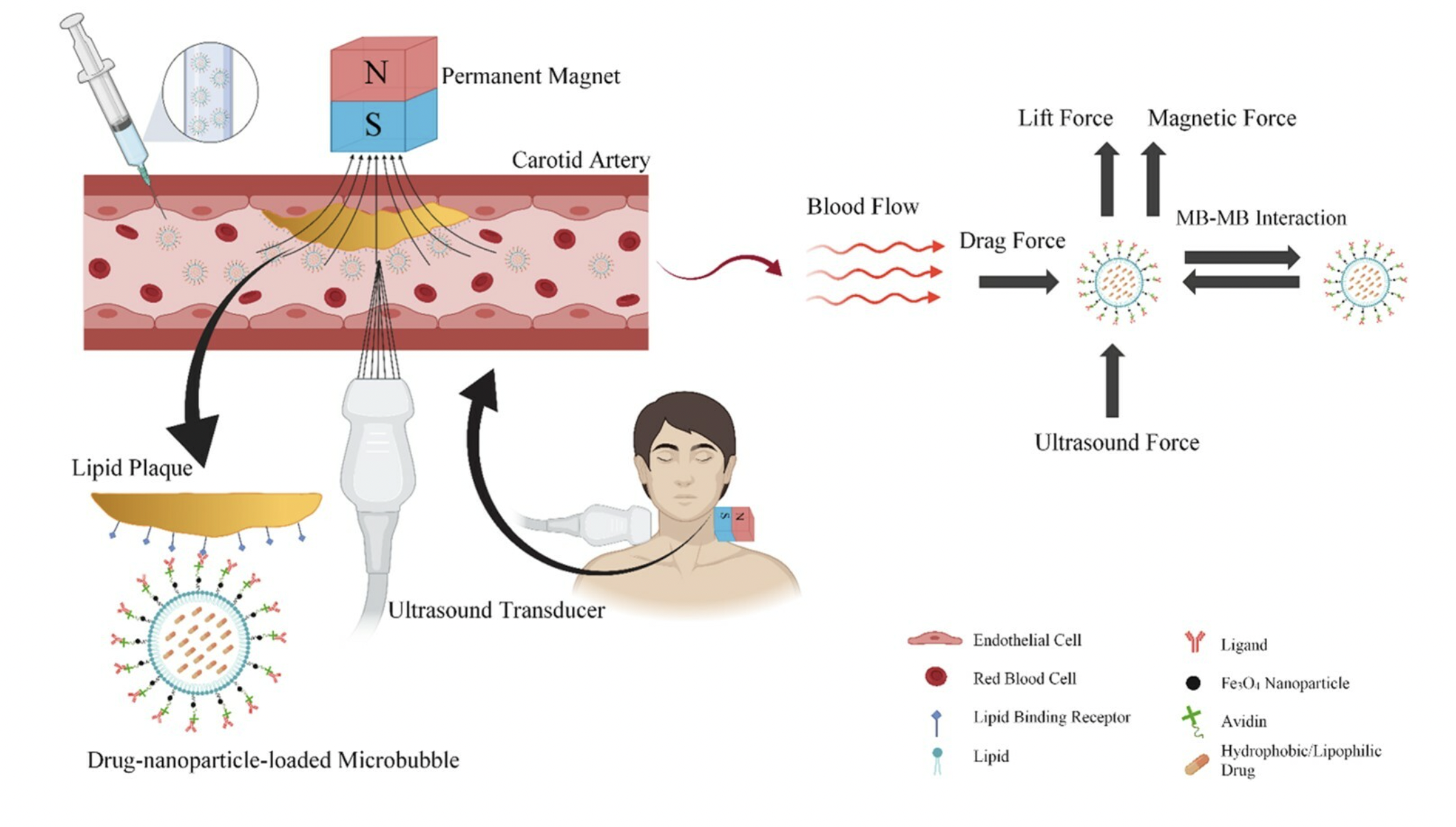 |
Drug delivery and adhesion of magnetic nanoparticles coated nanoliposomes and microbubbles to atherosclerotic plaques under magnetic and ultrasound fields Alishiri M, Ebrahimi S, Shamloo A, Boroumand A, Mofrad MRK Engineering Applications of Computational Fluid Mechanics, 15(1):1703-725, 2021 (KEYWORDS: RNA, DNA, Proteins) |
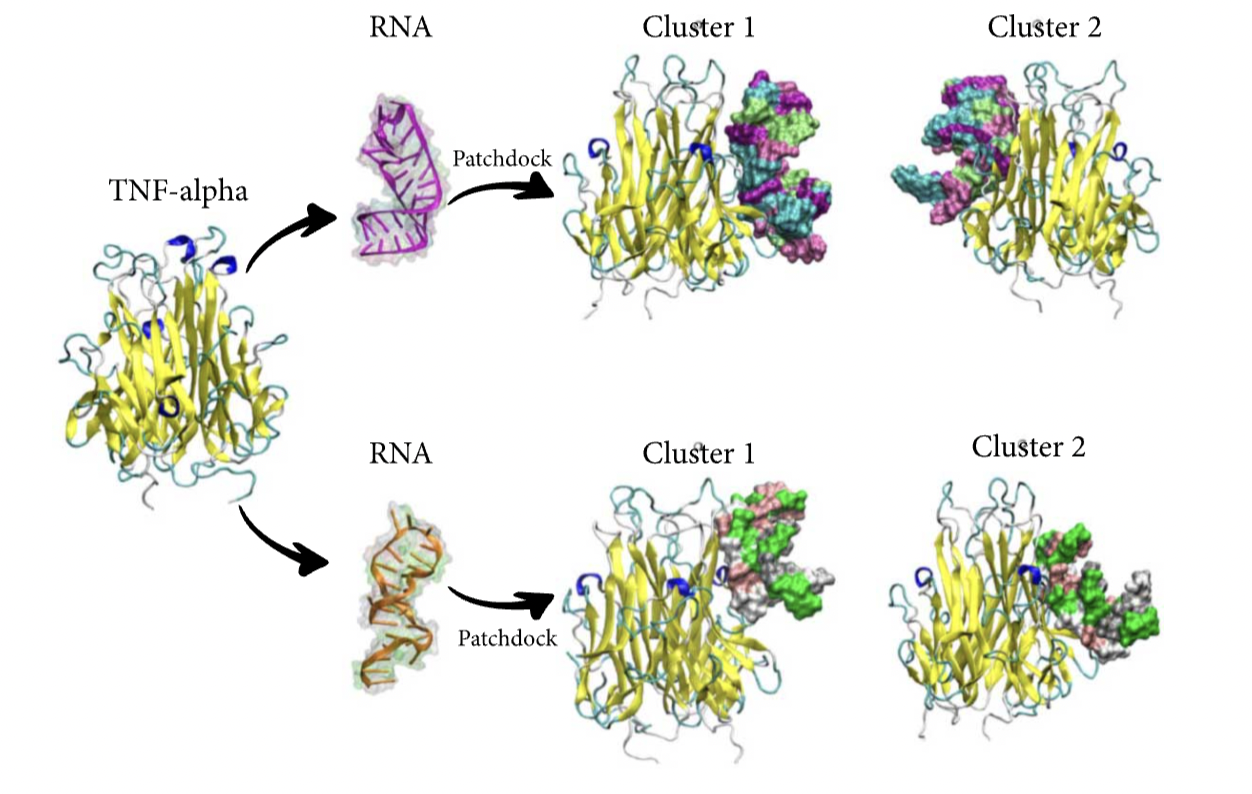 |
Atomic Scale Interactions between RNA and DNA Aptamers with the TNF-α Protein Asadzadeh H, Moosavi A, Alexandrakis G, Mofrad MRK BioMed Research International (Special Issue on Non-coding RNA in Cardiovascular Disease), 2021 (KEYWORDS: Atherosclerosis) |
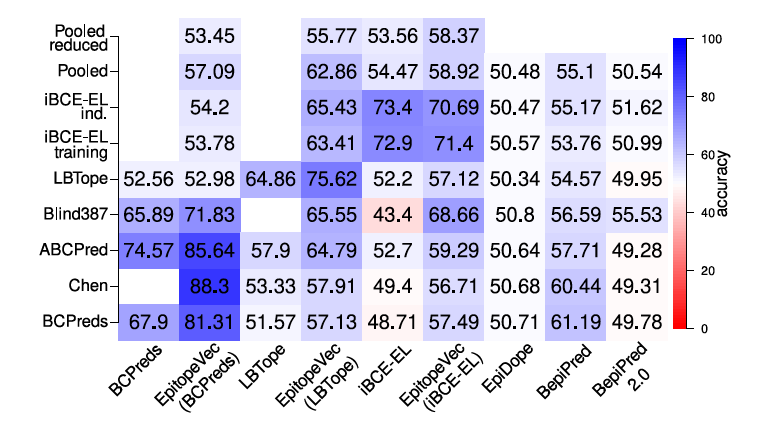 |
EpitopeVec: Linear Epitope Prediction Using Deep Protein Sequence Embeddings. Bahai A, Asgari E, Mofrad MRK, Kloetgen A, McHardy AC. Bioinformatics:btab467. doi: 10.1093/bioinformatics/btab467. (KEYWORDS: Deep Proteomics, ProtVec) |
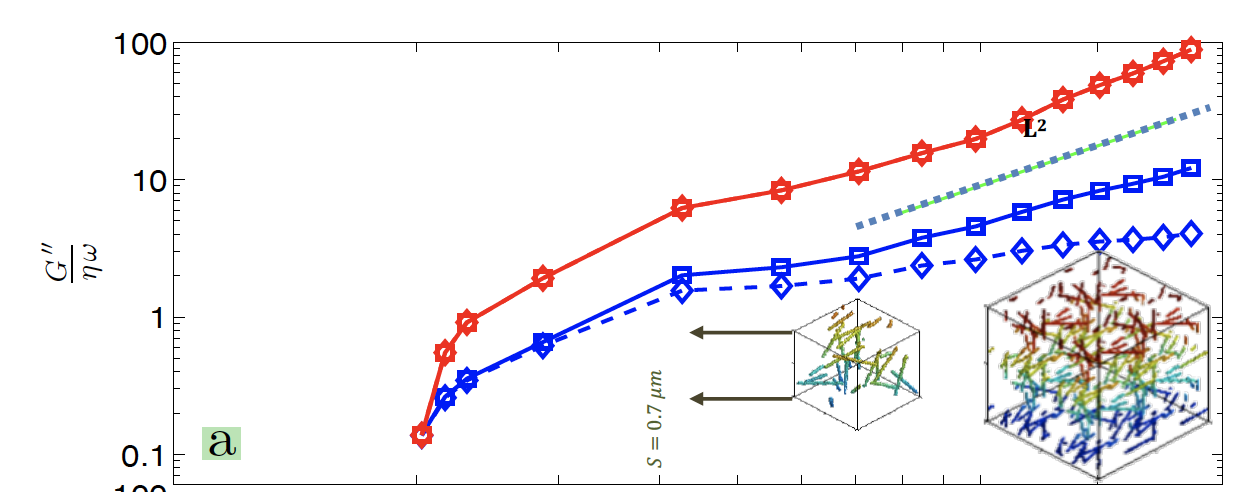 |
Hydrodynamic interactions significantly alter the dynamics of actin networks and result in a length scale dependent loss modulus. R Karimi, MR Alam, MRK Mofrad Journal of Biomechanics 120, 110352 (KEYWORDS: Cytoskeleton, Cell mechanics, Cytoskeletal Rheology) |
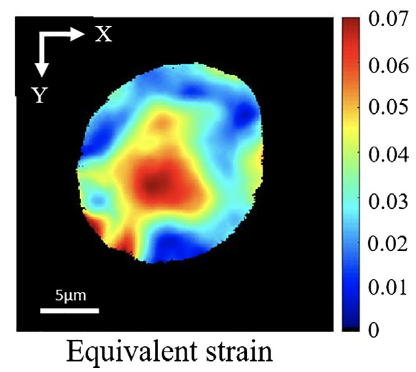 |
Intranuclear strain in living cells subjected to substrate stretching: A combined experimental and computational study. S Tsukamoto, T Asakawa, S Kimura, N Takesue, MRK Mofrad, N Sakamoto Journal of Biomechanics 119, 110292. (KEYWORDS: Cytoskeleton, Cell mechanics, Cytoskeletal Rheology) |
 |
PFP-WGAN: Protein function prediction by discovering Gene Ontology term correlations with generative adversarial networks. SF Seyyedsalehi, M Soleymani, HR Rabiee, MRK Mofrad PLoS One 16 (2), e0244430. (KEYWORDS: Cytoskeleton, Cell mechanics, Cytoskeletal Rheology) |
 |
Strain-stiffening and strain-softening responses in random viscoelastic fibrous networks: interplay between fiber orientation and viscoelastic softening. N Zolfaghari, M Moghimi Zand, MRK Mofrad Soft Materials 18 (4), 373-385. (KEYWORDS: Cytoskeleton, Cell mechanics, Cytoskeletal Rheology) |
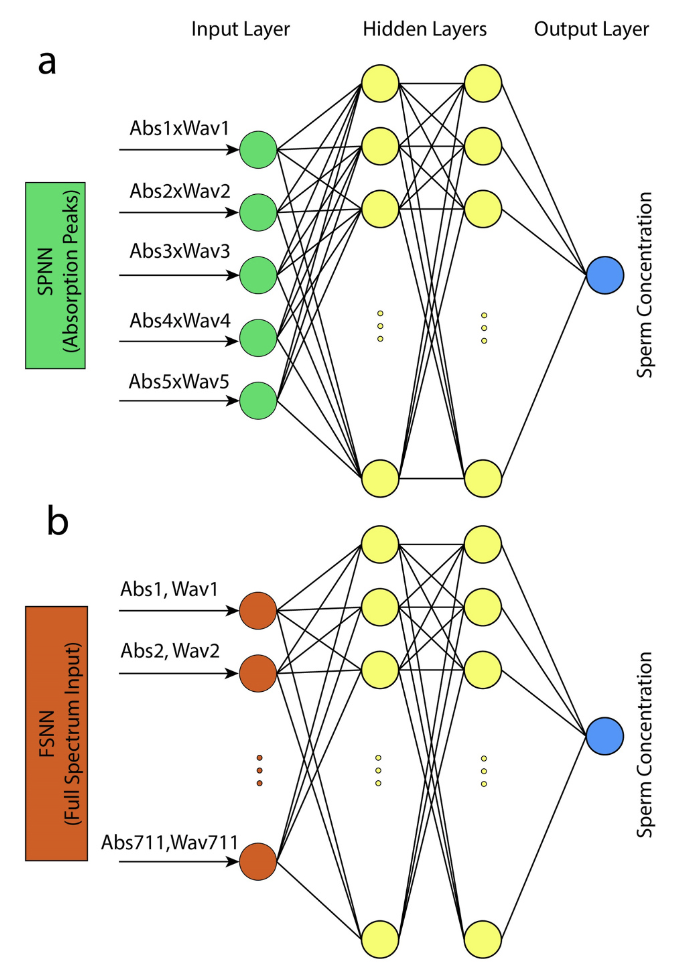 |
Quantification of human sperm concentration using machine learning-based spectrophotometry. A Lesani, S Kazemnejad, MM Zand, M Azadi, H Jafari, MRK Mofrad, R. Nosrati Computers in Biology and Medicine 127, 104061 (KEYWORDS: Machine Learning Application) |
 |
Machine learning for endoleak detection after endovascular aortic repair Talebi S*, Madani MH*, Madani A, Chien A, Shen J, Mastrodicasa D, Fleischmann D, Chan FP*, Mofrad MRK*. Scientific Reports. 2020DOI: 10.1038/s41598-020-74936-7 (KEYWORDS: Cardiovascular disease, machine learning.) |
 |
A splice acceptor variant in HLA-DRA affects the conformation and cellular localization of the class II DR alpha-chain Didonna A, Damotte V, Shams H, Matsunaga A, Caillier S, Dandekar R, Misra M, Mofrad M, Oksenberg J, Hollenbach JImmunology. 2020 DOI: https://doi.org/10.1111/IMM.13273 (KEYWORDS: neurodegenerative disease, human leukocyte antigen, immune response, antigen presentation, protein folding.) |
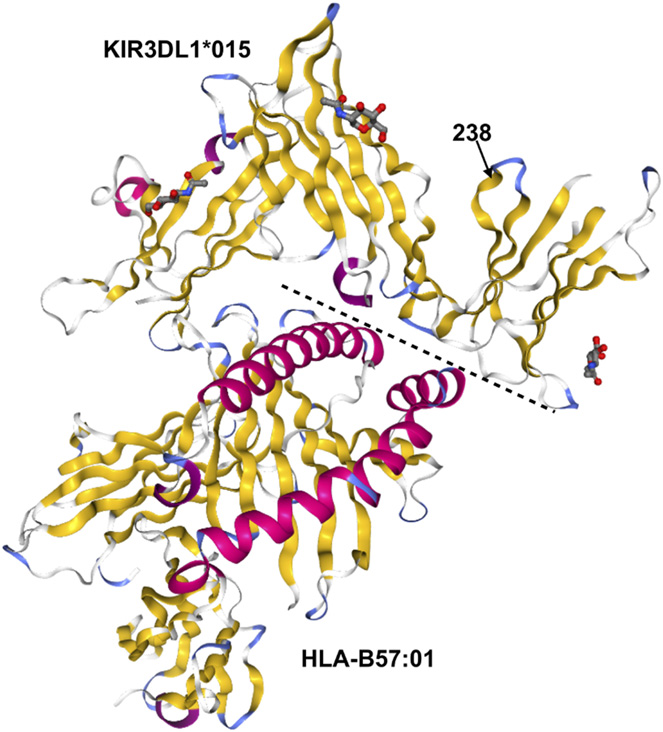 |
Killer Cell Immunoglobulin-like Receptor Variants Are Associated with Protection from Symptoms Associated with More Severe Course in Parkinson Disease Anderson KM, Augusto DG, Dandekar R, Shams H, Zhao C, Yusufali T, Montero-Martın G, Marin WM, Nemat-Gorgani N, Creary LE, Caillier L, Mofrad MRK, Parham P, Fernandez-Vina M, Oksenberg JR, Norman PJ, Hollenbach JA Journal of Immunology. 2020 DOI: 10.4049/jimmunol.2000144.(KEYWORDS: neurodegenerative disease, Killer cell immunoglobulin-like receptor 3DL1 protein) |
 |
Nanoscale integrin cluster dynamics controls cellular mechanosensing via FAK Y397 phosphorylation. Cheng B, Wan W, Huang G, Li Y, Genin GM, Mofrad MRK, Lu TJ, Xu F, Lin M. Science Advance. 6(10):eaax1909 DOI: 10.1126/sciadv.aax19092020.(KEYWORDS: Cellular Mechanotransduction, Focal Adhesions, FAK, Integrin Clustering) |
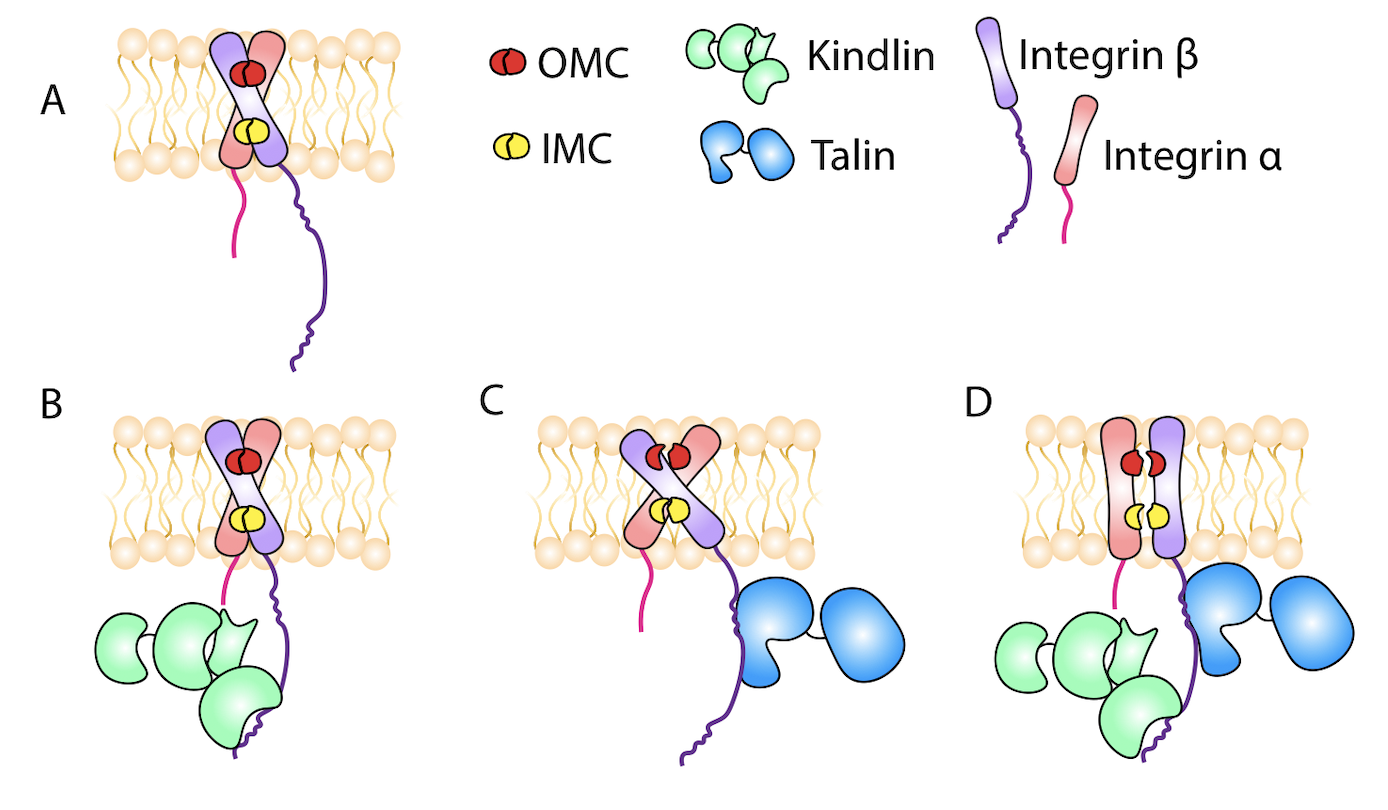 |
Kindlin Assists Talin to Promote Integrin Activation Haydari Z, Shams H, Jahed Z, Mofrad MRK. Biophysical Journal. 118(8):1977-1991, APRIL 21, 2020(KEYWORDS: Cellular Mechanotransduction, Focal Adhesions, FAK, Integrin Activation, Kindlin, Talin) |
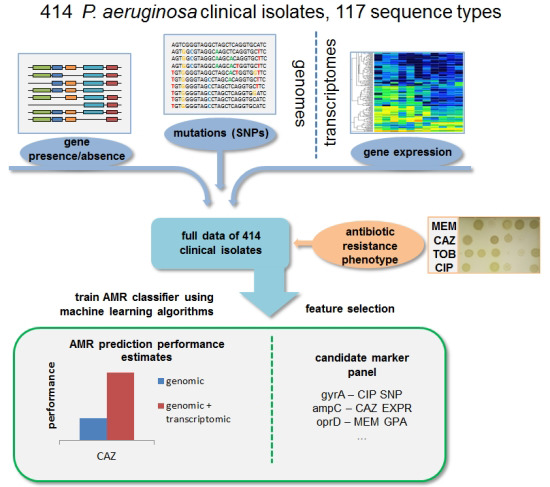 |
Predicting antimicrobial resistance in Pseudomonas aeruginosa with machine learning‐enabled molecular diagnostics. Khaledi A, Weimann A, Schniederjans M, Asgari E, Kuo TH, Oliver A, Cabot G, Kola A, Gastmeier P, Hogardt M, Jonas D, Mofrad MRK, Bremges A, McHardy AC, Häussler S. EMBO Molecular Medicine. e10264, 2020.(KEYWORDS: Bioinformatics, Machine Learning) |
 |
The CAFA challenge reports improved protein function prediction and new functional annotations for hundreds of genes through experimental screens. Zhou N et al. Genome Biology, 20:244, 2019(KEYWORDS: Proteomics, Bioinformatics, protein function prediction) |
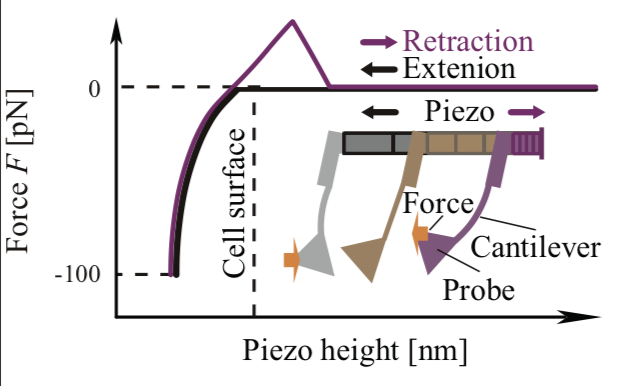 |
Talin is required to increase stiffness of focal molecular complex in its early formation process. Nakao N, Maki K, Mofrad MRK, Adachi T Biochemical and Biophysical Research Communications, 518 (3):579-583, 2019.(KEYWORDS: Focal Adhesions, Talin, Mechanotransduction) |
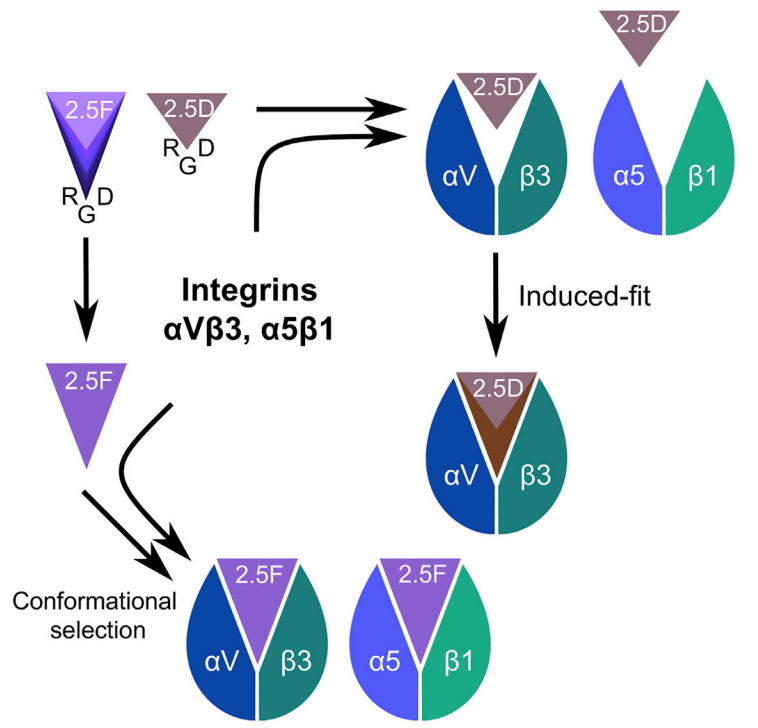 |
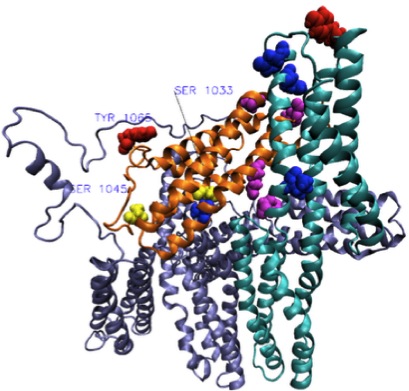
Structural Basis of the Differential Binding of Engineered Knottins to Integrins αVβ3 and α5β1. Van Agthoven JF, Shams H et al. Structure, 27, 1443–1451, 2019.(KEYWORDS: Integrins, αVβ3, α5β1) |
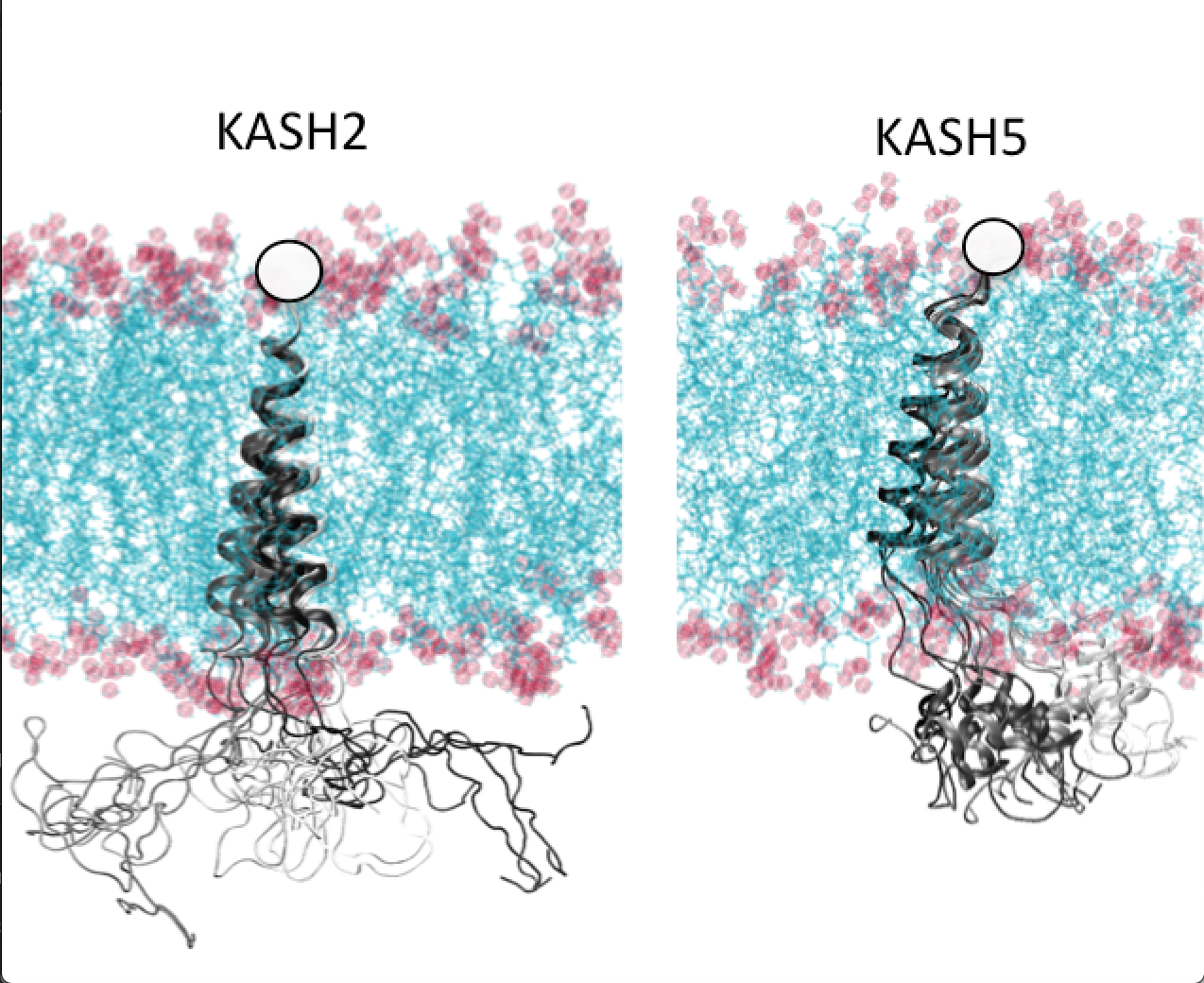 |
Role of KASH domain lengths in the regulation of LINC complexes Jahed Z, Hao H, Thakkar V, Vu UT, Valdez VA, Rathish A, Tolentino C, Kim SJC, Fadavi D, Starr DA, Mofrad MRK. Molecular Biology of the Cell (MBoC), 30(16):2076-2086.(KEYWORDS: LINC Complex, SUN/KASH, Nuclear Mechanics, Mechanotransduction) |
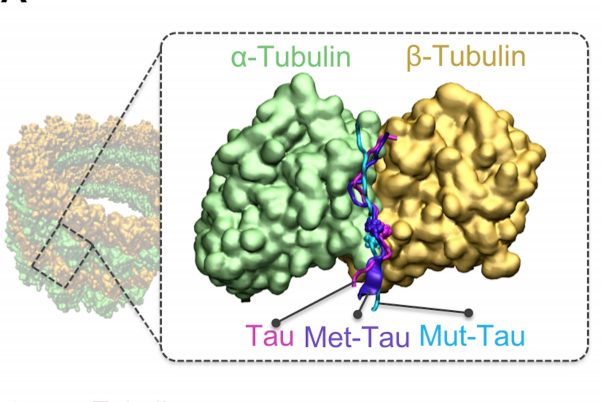 |
Sex-specific Tau methylation patterns and synaptic transcriptional alterations are associated with neural vulnerability during chronic neuroinflammation Didonna A, Cantó E, Shams H, Isobe N, Zhao C, Caillier SJ, Condello C, Yamate-Morgan H, Tiwari-Woodruff SK, Mofrad MRK, Hauser SL, Oksenberg JR Journal of Autoimmunity. 101:56-692, 2019.(KEYWORDS: Tau protein, multiple sclerosis, post-translational modifications, neuroinflammation) |
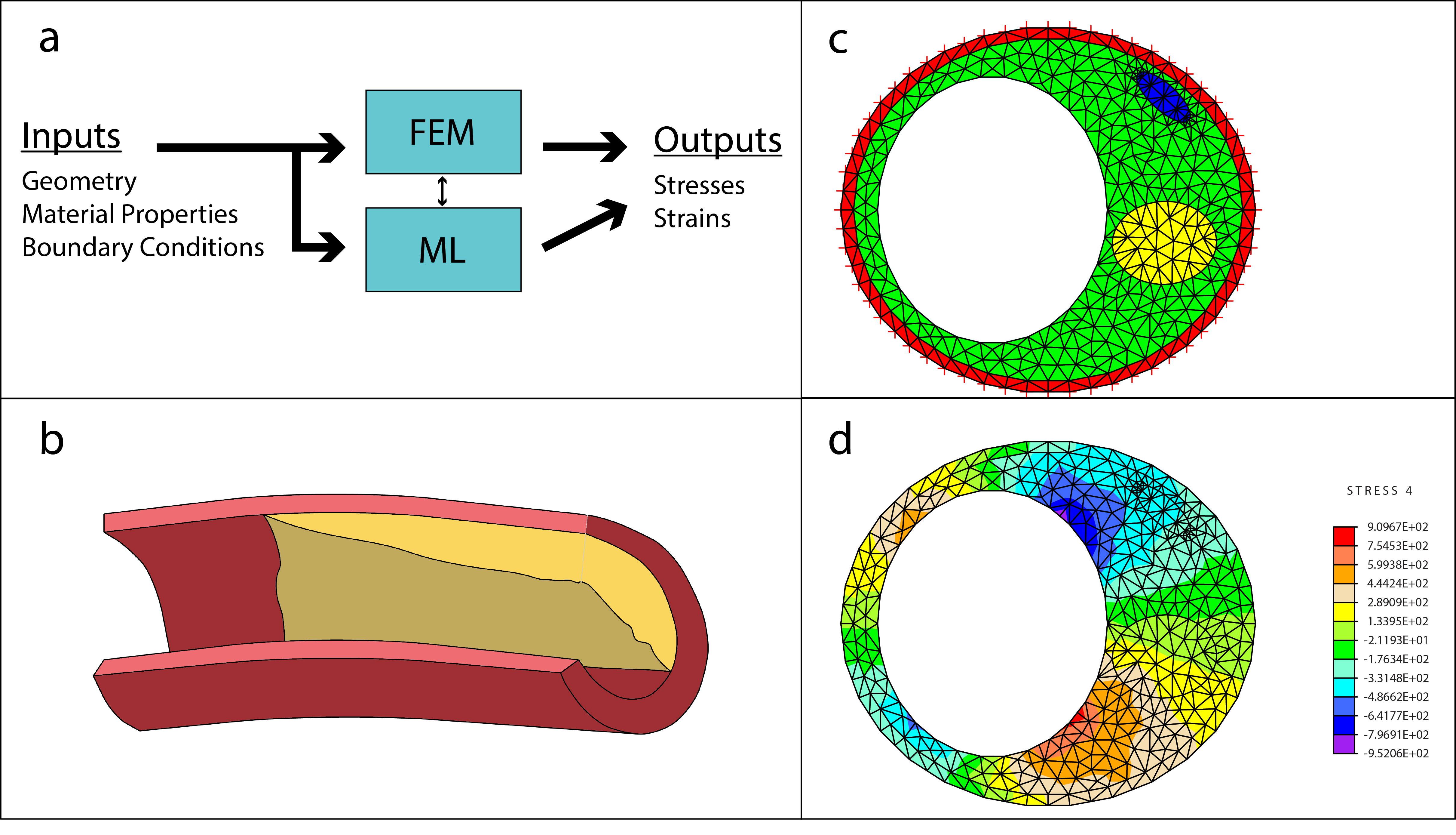 |
Bridging finite element and machine learning modeling: stress prediction of arterial walls in atherosclerosis Madani A, Bakhaty A, Kim J, Mubarak Y, Mofrad MRK. ASME J. Biomechanical Engineering. 141(8): 084502, 2019(KEYWORDS: Machine Learning, Deep Learning, Atherosclerois, Finite Element Modeling) |
 |
Kindlin is mechanosensitive: A force-induced conformational switch mediates intracellular crosstalk among integrins Jahed Z, Haydari Z, Mofrad MRK. Biophysical Journal. 116(6): 1011-24, 2019.(KEYWORDS: Cellular mechanotransduction, Focal adhesions, kindlin, integrin) |
 |
The Nucleus Feels the Force, LINCed In or Not! Jahed Z, Mofrad MRK. Current Opinion in Cell Biology. Volume 58, June 2019, Pages 114-119(KEYWORDS: Nucleus, Mechanotransduction, Nuclear envelop, LINC complex, SUN/KASH) |
 |
Probabilistic variable-length segmentation of protein sequences for discriminative motif discovery (DiMotif) and sequence embedding (ProtVecX). Asgari E, McHardy AC, Mofrad MRK. Scientific Reports. (2019) 9:3577 doi:10.1038/s41598-019-38746-w(KEYWORDS: Deep Learning, Protein Bioinformatics, Deep Proteomics, ProtVec) |
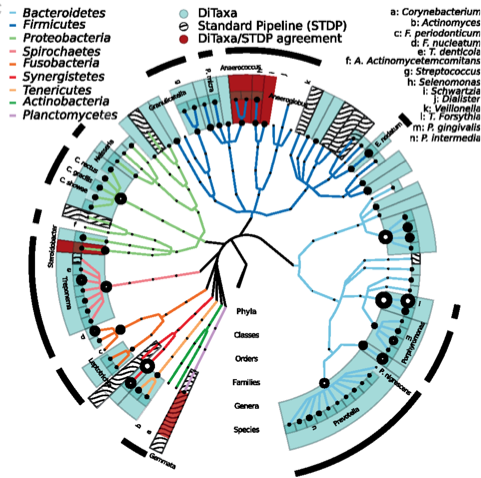 |
DiTaxa: Nucleotide-pair encoding of 16S rRNA for host phenotype and biomarker detection. Asgari E, Münch PC, Lesker TR, McHardy AC, Mofrad MRK. Bioinformatics. 2018 Nov 30. doi: 10.1093/bioinformatics/bty954. (KEYWORDS: Deep Learning, Microbiome, 16S rRNA data analysis) |
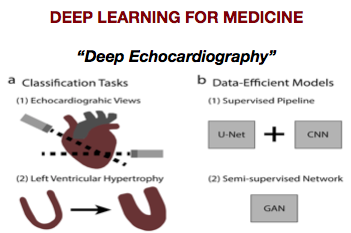 |
Deep echocardiography: data-efficient supervised and semi-supervised deep learning towards automated diagnosis of cardiac disease Madani A, Rui Ong J, Tibrewal A, Mofrad MRK. npj Digital Medicine. 1: 59 (2018)(KEYWORDS: Deep Learning, Machine Learning, Medicine, Cardiac Disease) |
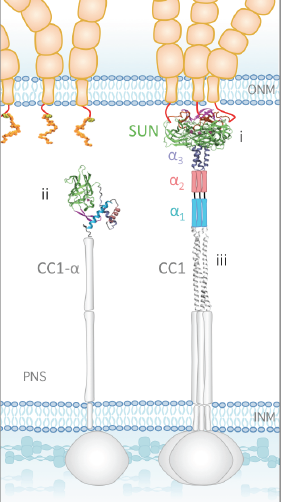 |
A molecular model for LINC complex regulation: Activation of SUN2 for KASH binding Jahed Z, Vu UT, Fadavi D, Ke H, Rathish A, Kim SCJ, Feng W, Mofrad MRK. Molecular Biology of the Cell (MBoC), 29(16):2012-2023, 2018.(KEYWORDS: LINC Complex, SUN/KASH, Nuclear Mechanics, Mechanotransduction) |
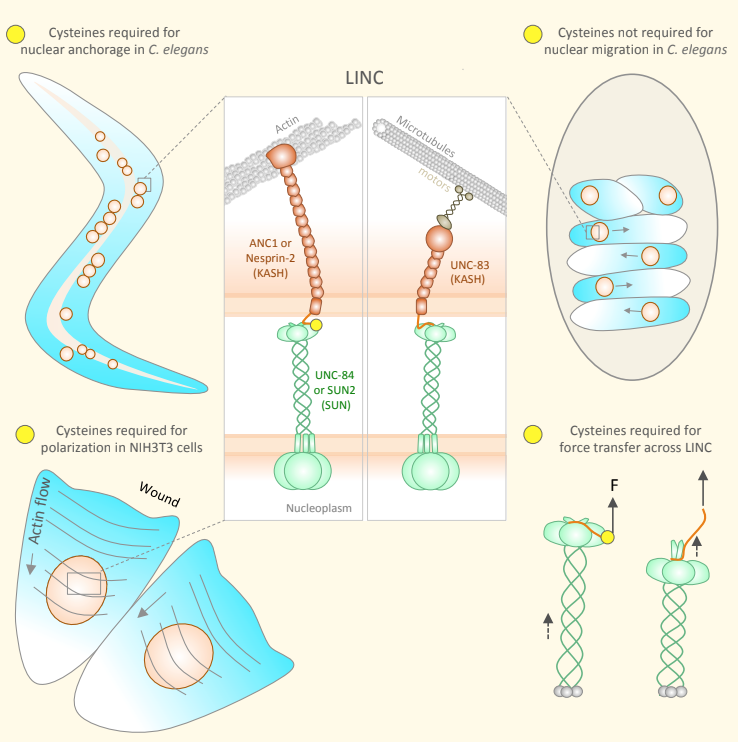 |
Conserved SUN-KASH interfaces mediate LINC complex-dependent nuclear movement and positioning Cain NC, Jahed Z, Schoenhofen A, Valdez VA, Elkin B,Hao H, Harris NJ, Herrera LA, Woolums BW, Mofrad MRK, Luxton G, Starr DA Current Biology, 28:3086–3097, October 8, 2018.(KEYWORDS: LINC Complex, SUN/KASH, Nuclear Mechanics, Mechanotransduction) |
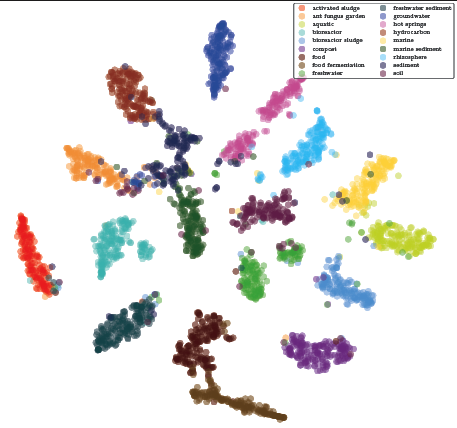 |
MicroPheno: Predicting environments and host phenotypes from 16S rRNA gene sequencing using a k-mer based representation of shallow sub-samples Asgari E, Garakani K, McHardy AC, Mofrad MRK. Bioinformatics, Volume 34, Issue 13, 1 July 2018, Pages i32–i42(KEYWORDS: Microbiome, Phenotypes, Genomics, Bioinformatics, Deep Learning, Machine Learning) |
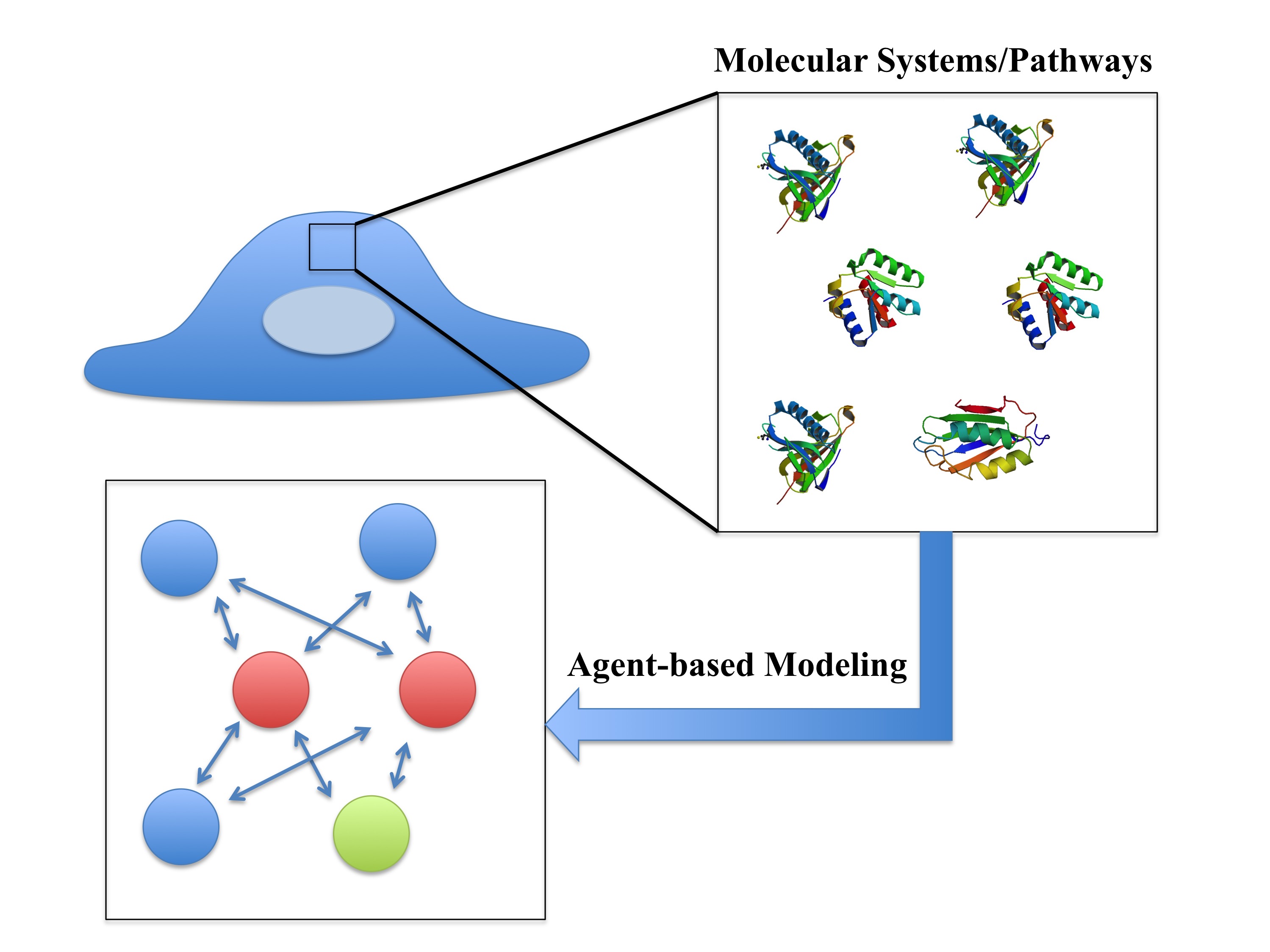 |
Agent-based Modeling in Molecular Systems Biology Soheilypour M, Mofrad MRK. BioEssays, 2018 Jul;40(7):e1800020. doi: 10.1002/bies.201800020Chosen for BioEssays Issue Highlights(KEYWORDS: Molecular systems biology, Complex systems, Computational biology) |
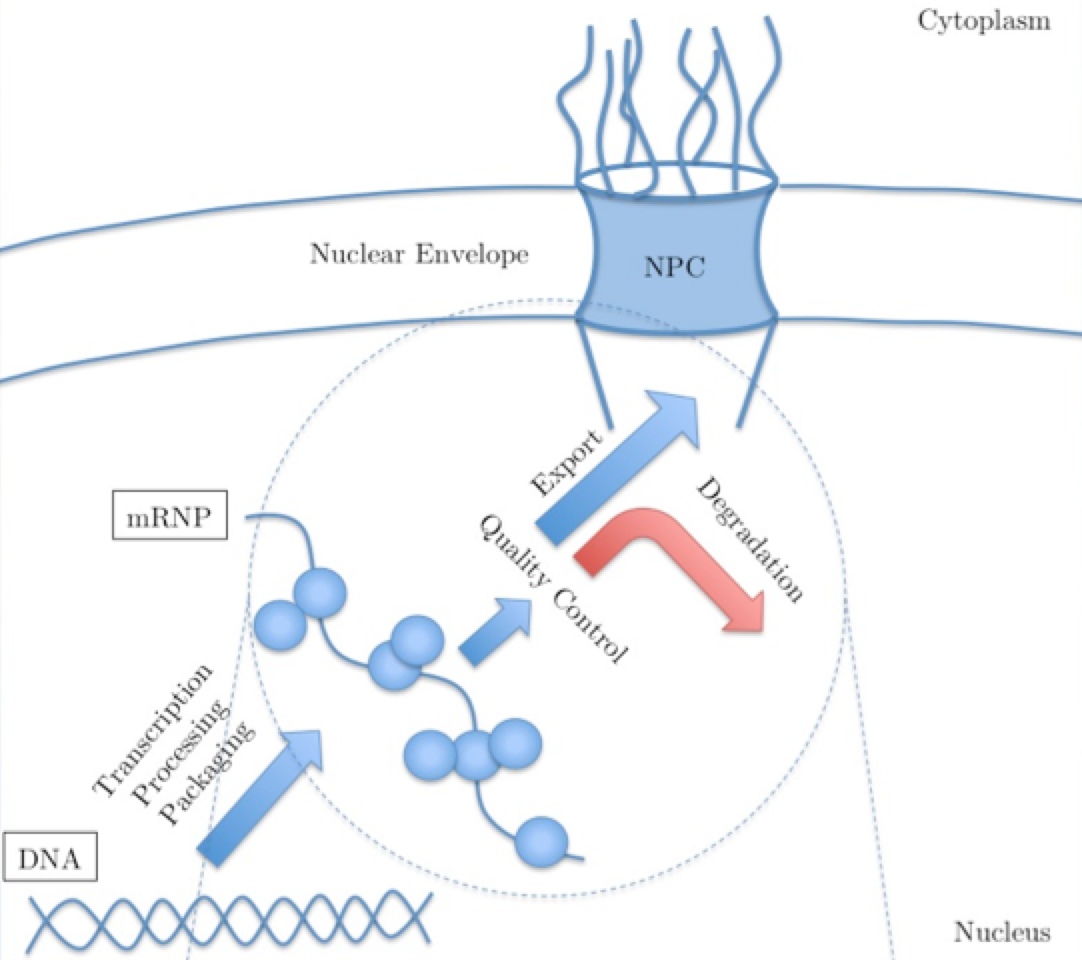 |
Quality Control of mRNAs at the Entry of the Nuclear Pore: Cooperation in a Complex Molecular System Soheilypour M, Mofrad MRK. Nucleus, Volume 9, Issue 1, Pages 202-211.(KEYWORDS: Nuclear Pore Complex, mRNA Export, Mechanotransduction) |
 |
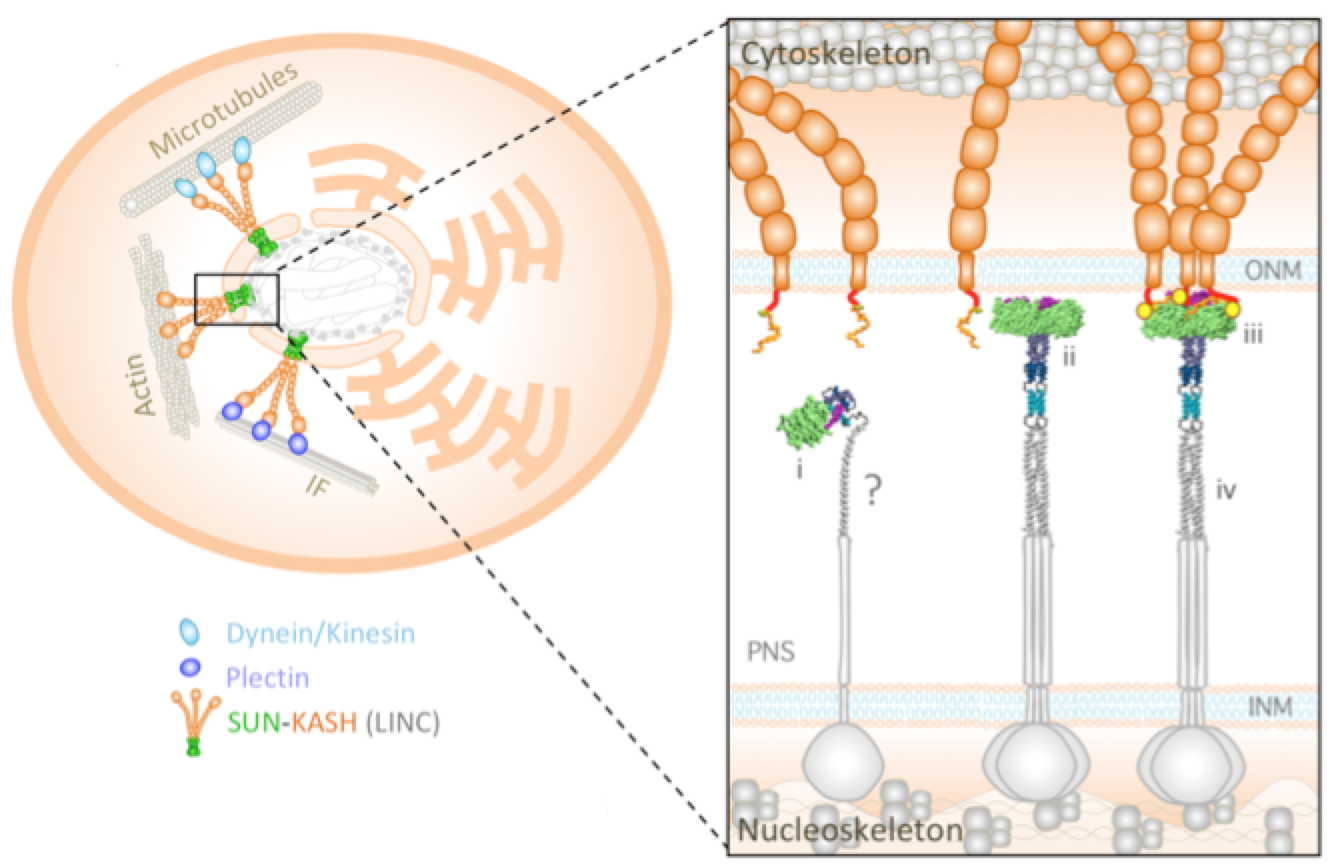
Molecular insights into the mechanisms of SUN1 oligomerization in the nuclear envelope Jahed Z, Fadavi D, Vu UT, Asgari E, Luxton GWG, Mofrad MRK. Biophysical Journal, Volume 114, Issue 5, p1190–1203, 13 March 2018.(KEYWORDS: LINC Complex, Nuclear Mechanics, Mechanotransduction) |
 |
Mechanical LINCs of the nuclear envelope: Where SUN meets KASH Jahed Z, Mofrad MRK. Extreme Mechanics Letters (special issue on Mechanobiology), Volume 20, April 2018, Pages 99-103.(KEYWORDS: LINC Complex, Nuclear Mechanics, Mechanotransduction) |
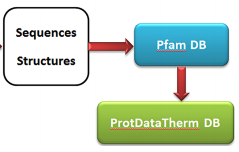 |
ProtDataTherm: A database for thermostability analysis and engineering of proteins Modarres HP, Mofrad MRK, Sanati-Nezhad A. PLoS One, 13(1): e0191222, 2018.(KEYWORDS: Bioinformatics, Protein Engineering) |
 |
The ‘stressful’ life of cell adhesion molecules: On the mechanosensitivity of integrin adhesome Shams H, Hoffman BD, Mofrad MRK. ASME Journal of Biomechanical Engineering, 2017 Dec 22. doi: 10.1115/1.4038812(KEYWORDS: Focal Adhesions, Mechanotransduction, Integrin, Vinculin, Talin) |
 |
Fast and accurate view classification of echocardiograms using deep learning Madani A, Arnaout R, Mofrad MRK*, Arnaout R*. npj Digital Medicine, In Press.(KEYWORDS: Deep neural networks, Machine learning, Echocardiography, Precision Medicine) |
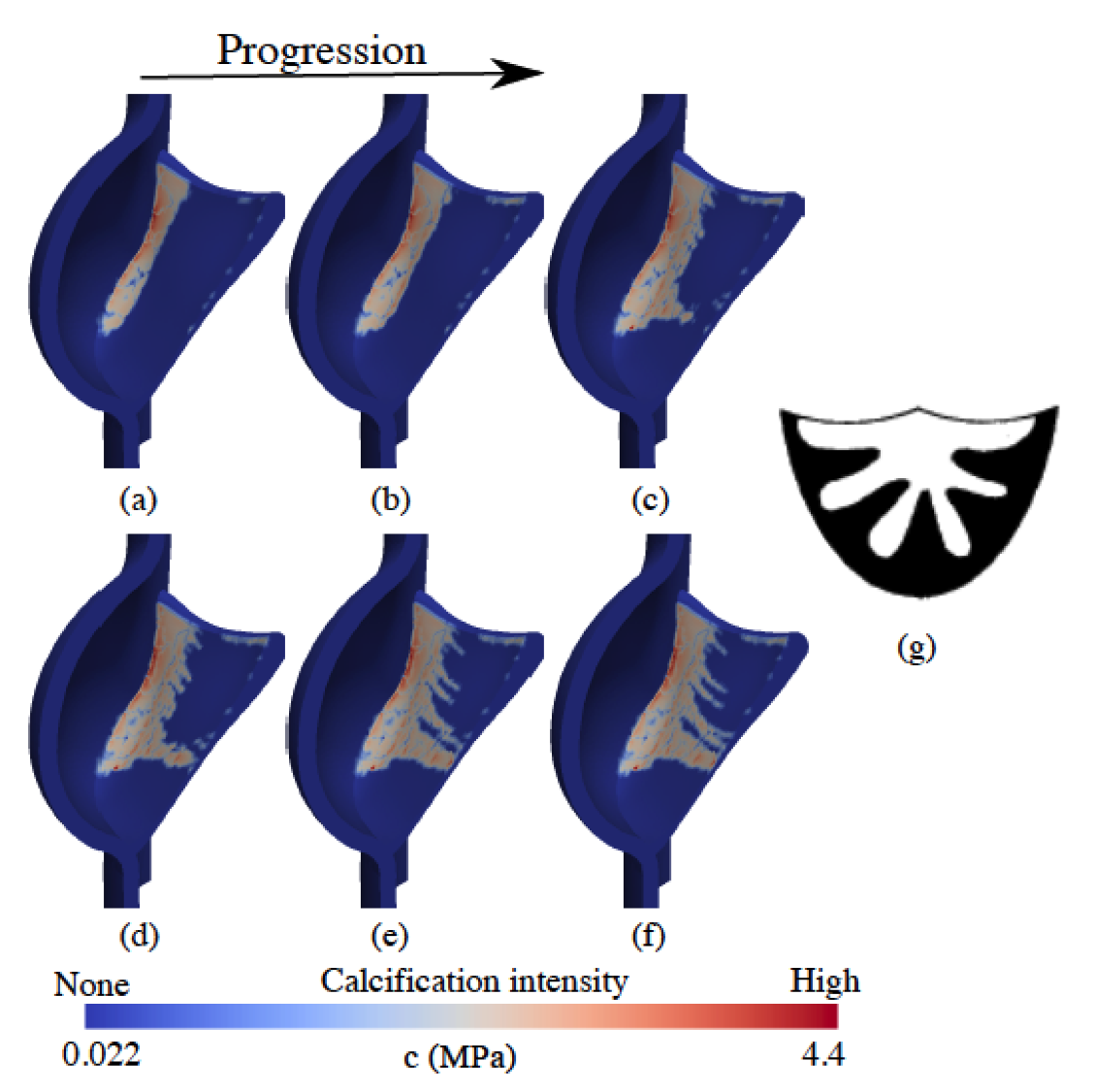 |
A strain-based finite element model for calcification progression in aortic valves Arzani A, Mofrad MRK. Journal of Biomechanics, 2017 Dec 8;65:216-220. (KEYWORDS: cardiovascular biomechanics, aortic valve) |
 |
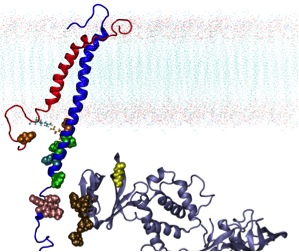
Interaction with α-actinin induces a structural kink in the transmembrane domain of β3-integrin and impairs signal transduction Shams H, Mofrad MRK. Biophysical Journal, 113(4):948–956, 2017. (KEYWORDS: mechanotransduction, focal adhesions) |
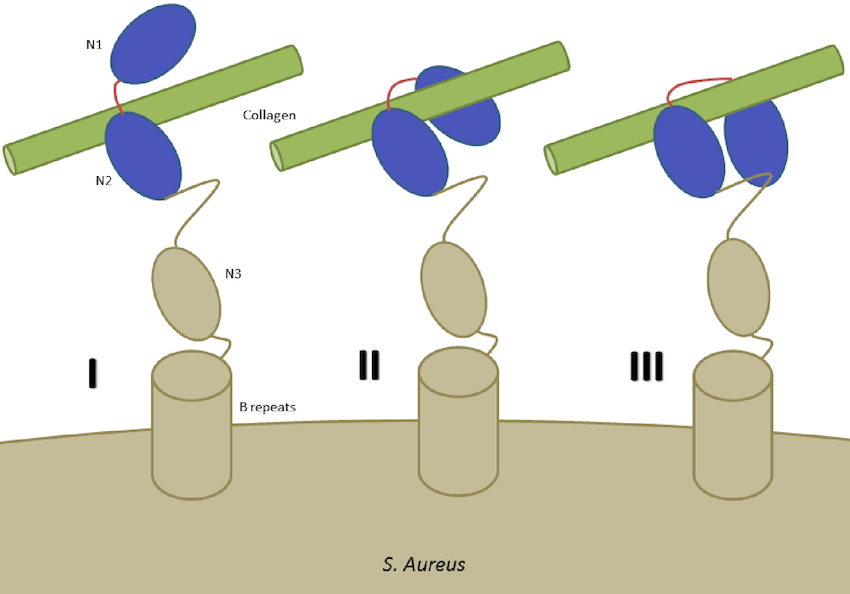 |
Molecular mechanics of staphylococcus aureus adhesin, CNA, and the inhibition of bacterial adhesion by stretching collagen Madani A, Garakani K, Mofrad MRK. PLoS One, 2017 Jun 30;12(6):e0179601. (KEYWORDS: bacterial mechanics) |
 |
Mechanical contact characteristics of PC3 human prostate cancer cell on complex shaped silicon micropillars. Seo BB, Jahed Z, Coggan JA, Chau YY, Rogowski JL, Gu FX, Wen W, Mofrad MRK, Tsui TY. Materials. 2017, 10(8), 892; doi:10.3390/ma10080892. (KEYWORDS: cellular mechanotransduction) |
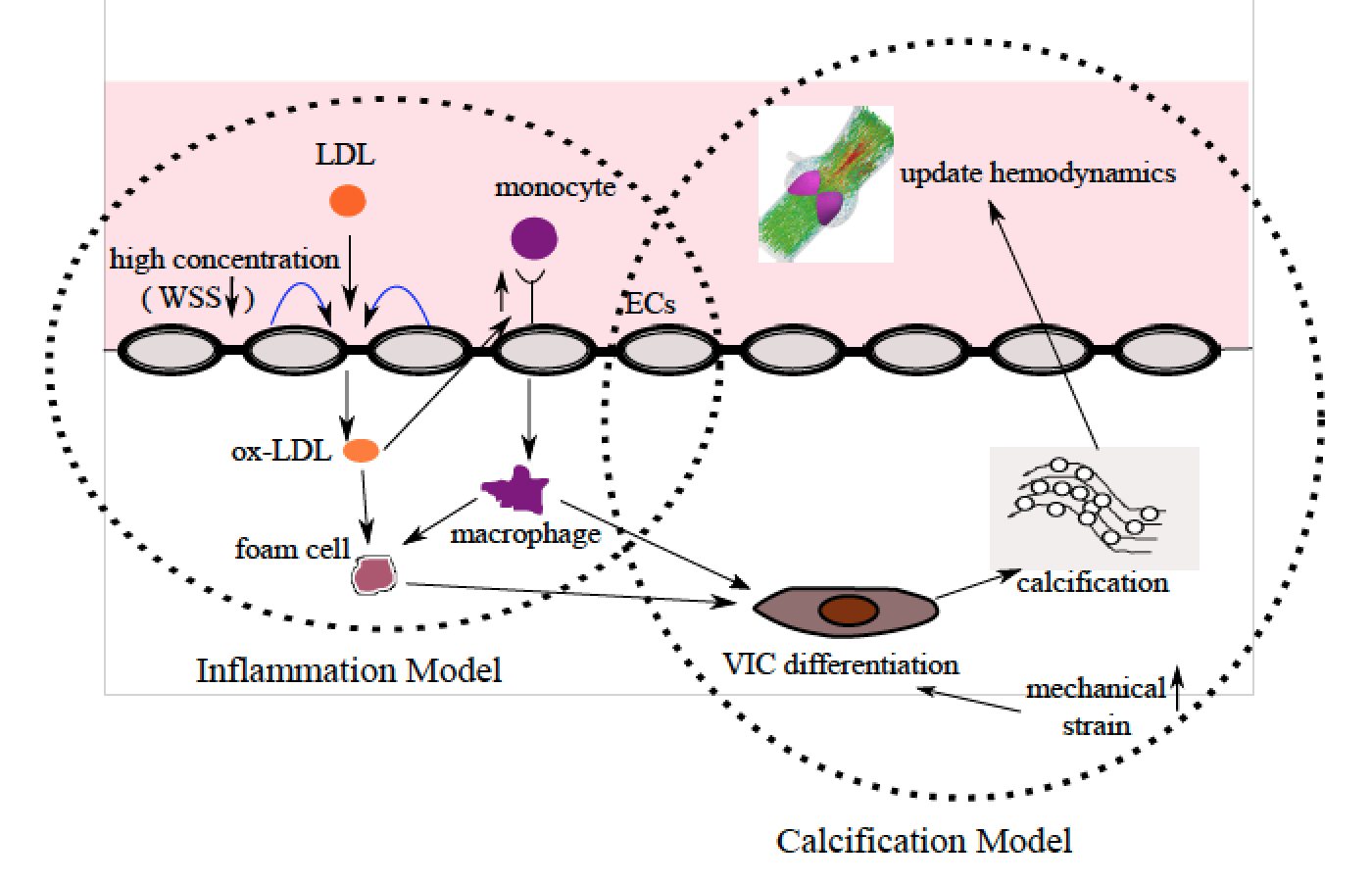 |
A multiscale systems biology model of calcific aortic valve disease progression Arzani A, Masters K, Mofrad MRK. ACS Biomaterials Science & Engineering, 2017, 3 (11), pp 2922–2933. (KEYWORDS: cardiovascular biomechanics, aortic valve) |
 |
Consistent Trilayer Biomechanical Modeling of the Aortic Valve Leaflet Tissue Bakhaty A, Govindjee S, , Mofrad MRK. Journal of Biomechanics, 61, 1-10. (KEYWORDS: cardiovascular biomechanics, aortic valve) |
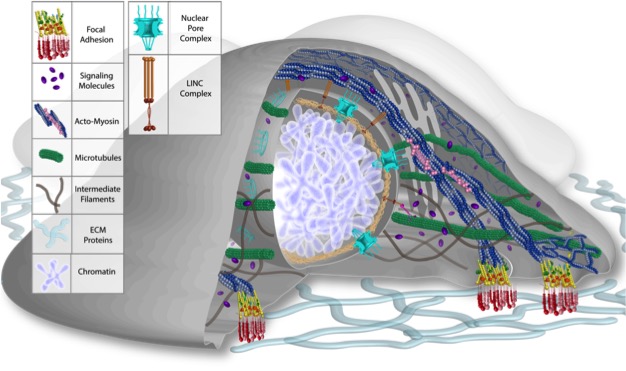 |
Looking “Under the Hood” of Cellular Mechanotransduction with Multiscale Computational Tools: A Systems Biomechanics Approach Shams H, Soheilypour M, Peyro M, Moussavi-Baygi R, Mofrad MRK. ACS Biomaterials Science & Engineering, 2017, 3 (11), pp 2712–2726. (KEYWORDS: cellular mechanotransduction, focal adhesions, nuclear pore complex, LINC complex) |
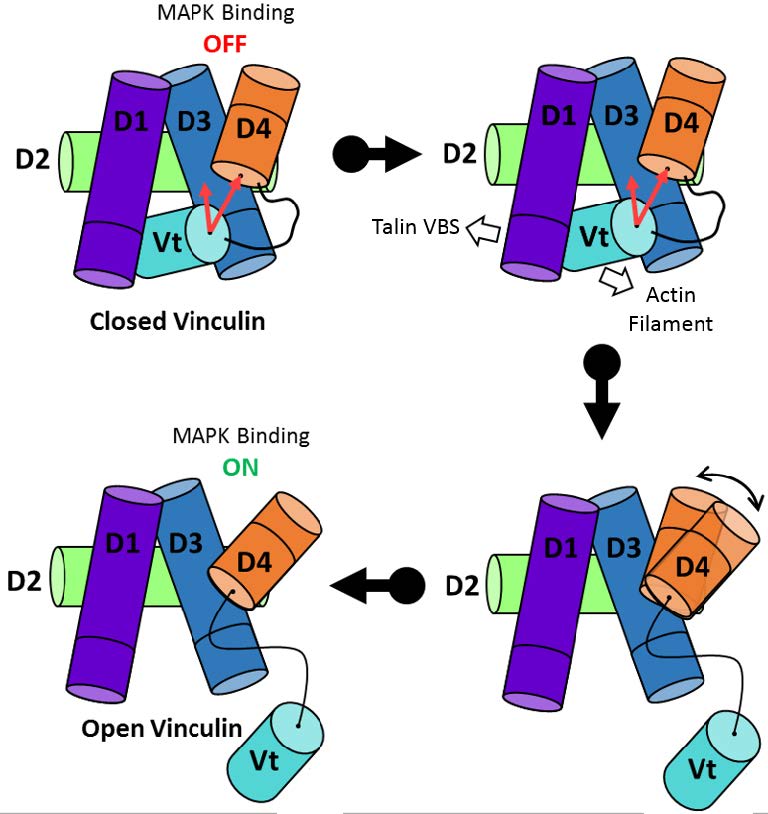 |
Mechanosensitive Conformation of Vinculin Regulates Its Binding to MAPK1 Garakani K, Shams H, Mofrad MRK. Biophysical Journal, 112, 1885–1893, May 9, 2017. (KEYWORDS: mechanotransduction, focal adhesions, vinculin, MAP Kinase) |
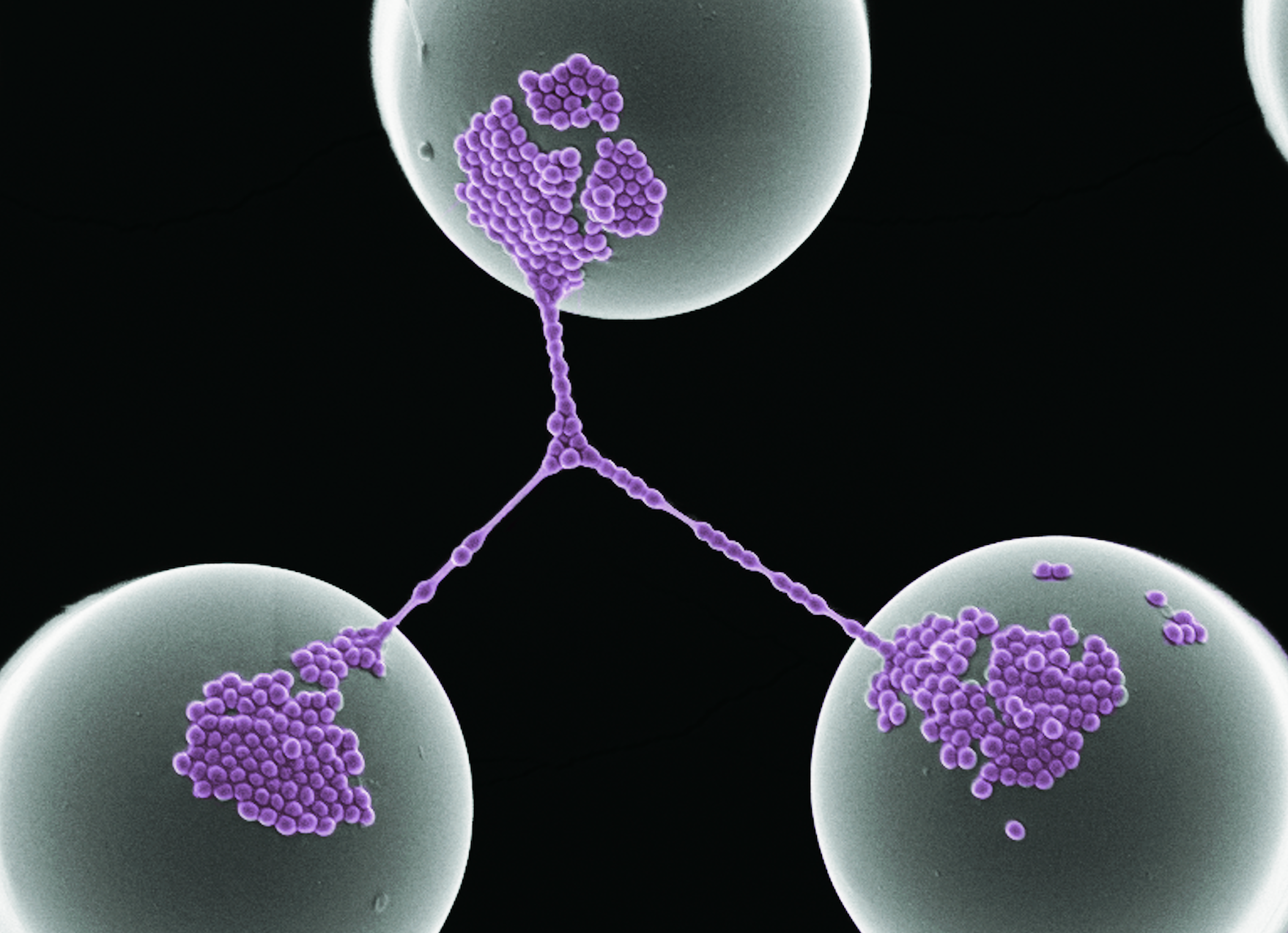 |
Bacterial Networks on Hydrophobic Micropillars Jahed Z, Shahsavan H, Verma MS, Rogowski JL, Seo BB, Zhao B, Tsui TY, Gu FX, Mofrad MRK. ACS Nano, 11 (1):675–683, 2017 doi: 10.1021/acsnano.6b06985. (KEYWORDS: mechanotransduction, bacterial adhesions, bacterial networks) |
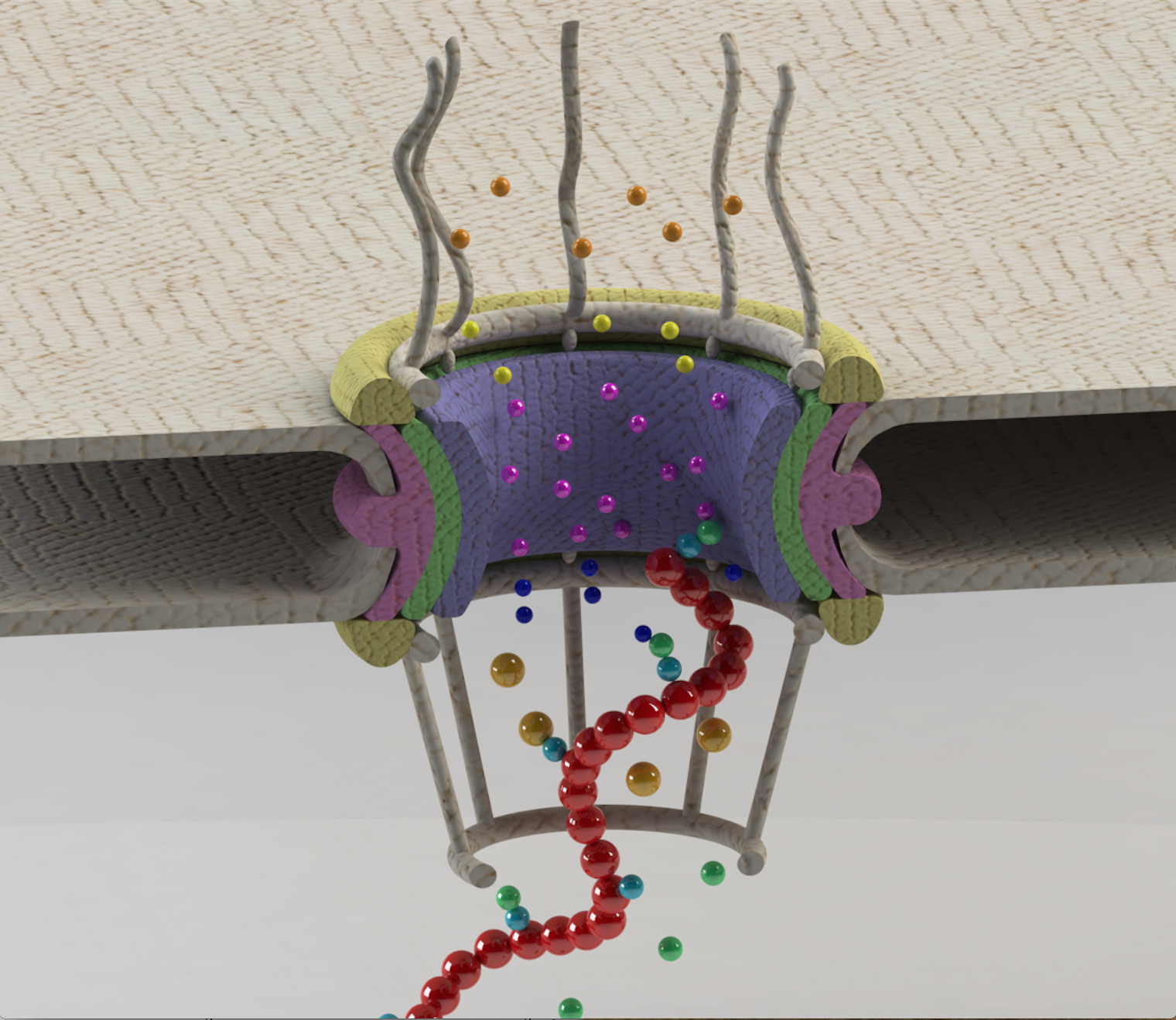 |
Regulation of RNA-binding proteins affinity to export receptors enables the nuclear basket proteins to distinguish and retain aberrant mRNAs Soheilypour M, Mofrad MRK. Scientific Reports, 6, Article number: 35380 (2016). |
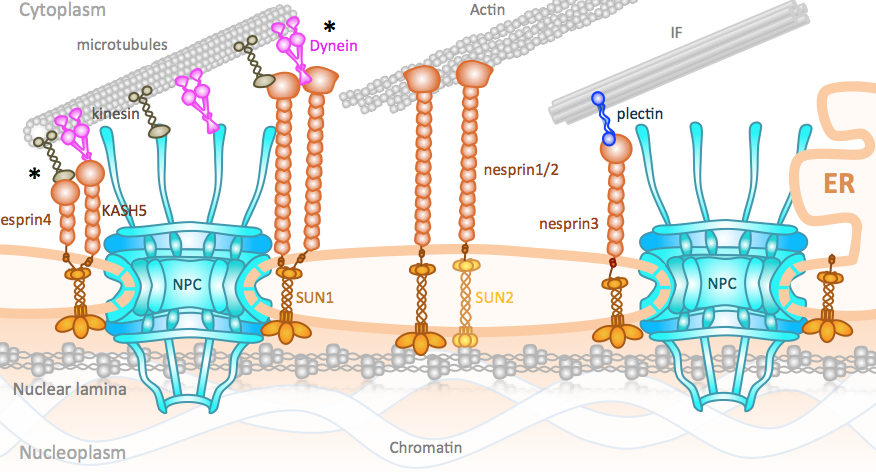 |
The LINC and NPC relationship: it’s complicated! Jahed Z, Soheilypour M, Peyro M, Mofrad MRK. Journal of Cell Science, J Cell Sci 129.17, 3219-3229 (2016). |
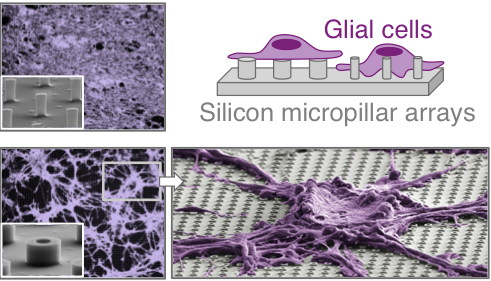 |
Differential Collective and Single Cell Behaviors on Silicon Micropillar Arrays Jahed Z, Zareian R, Chau Y, Seo B, West M, Tsui T, Wen W, Mofrad MRK. ACS Applied Materials & Interfaces. 8 (36):23604–23613, 2016. |
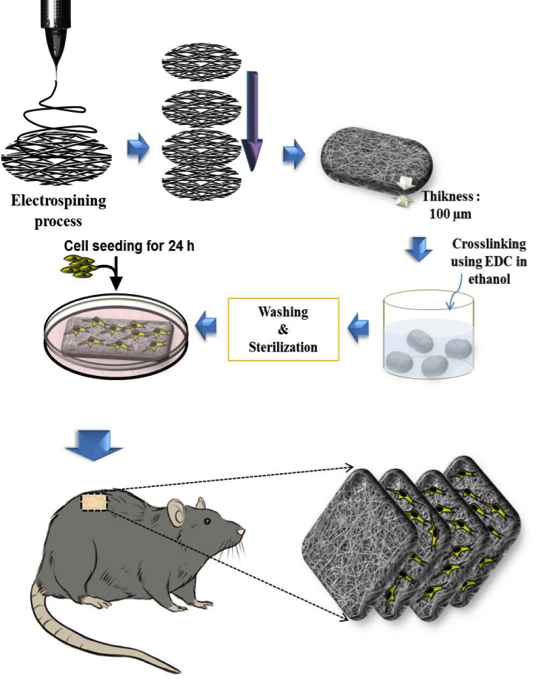 |
Gelatin/chondroitin sulfate nanofibrous scaffolds for stimulation of wound healing: In-vitro and in-vivo study Pezeshki-Modaress M, Mirzadeh H, Zandi M, Rajabi-Zeleti S, Sodeifi N, Aghdami N, Mofrad MRK. J Biomed Mater Res A. (2016). doi: 10.1002/jbm.a.35890. |
 |
Interferon Beta: From Molecular Level to Therapeutic Effects Haji Abdolvahab M, Mofrad MRK, Schellekens H. Int Rev Cell Mol Biol. (2016). 26:343-72. doi: 10.1016/bs.ircmb.2016.06.001. |
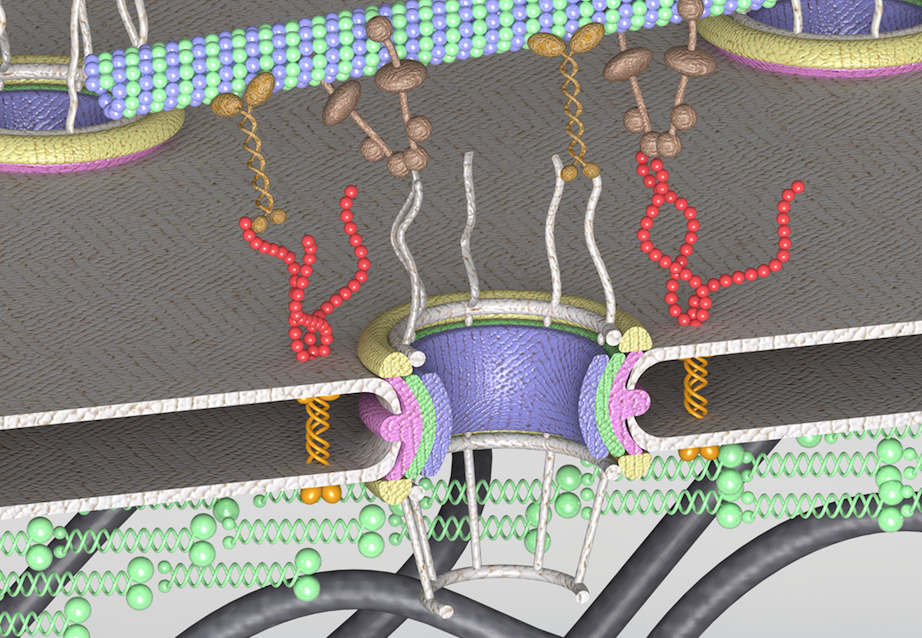 |
On the nuclear pore complex and its roles in nucleo-cytoskeletal coupling and mechanobiology Soheilypour M, Peyro M, Jahed Z, Mofrad MRK. Cellular and Molecular Bioengineering, DOI:10.1007/s12195-016-0443-x. |
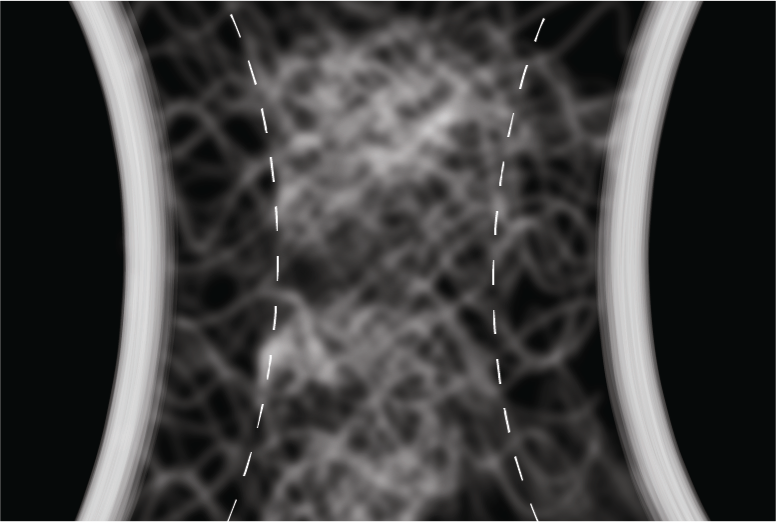 |
Rapid Brownian Motion Primes Ultrafast Reconstruction of Intrinsically Disordered Phe-Gly-Repeats Inside the Nuclear Pore Complex Moussavi-Baygi R, Mofrad MRK. Scientific Reports, 6, Article number: 29991 (2016). |
 |
Dynamic Regulation of α-Actinin’s Calponin Homology Domains on F-Actin Shams H, Golji J, Garakani K, Mofrad MRK. Biophysical Journal. 110, no. 6 (2016): 1444-1455. (KEYWORDS: mechanotransduction, focal adhesions, α-actinin, actin) |
 |
Protein thermostability engineering Modarres HP, Mofrad MRK, Sanati-Nezhad A. RSC Advances. 6, 115252-115270, 2016.(KEYWORDS: bioinformatics) |
 |
Micro and nanotechnologies in heart valve tissue engineering Hasan A, Saliba J, Pezeshgi-Modarres H, Bakhaty A, Nasajpour A, Mofrad MRK, Sanati-Nezhad A. Biomaterials 103 (2016) 278-292 (KEYWORDS: Heart Valves) |
 |
Enhanced intracellular delivery of small molecules and drugs via non-covalent ternary dispersions of single wall carbon nanotubes Boyer PD, Shams H, Baker SL, Mofrad MRK, Islam MF, Dahl KN. Journal of Materials Chemistry B. 4, 1324-1330, 2016 |
 |
Coupled Simulation of Heart Valves: Applications to Clinical Practice Bakhaty A, Mofrad MRK. Annals of Biomedical Engineering, 43(7)1626–1639, 2015. (KEYWORDS: cardiovascular biomechanics, aortic valve) |
 |
Nucleoporin’s Like Charge Regions Are Major Regulators of FG Coverage and Dynamics Inside the Nuclear Pore Complex Peyro M, Soheilypour M, Ghavami A, Mofrad MRK. PLoS ONE 10(12): e0143745. doi:10.1371/journal.pone.0143745, 2015 |
 |
Evolutionarily Conserved Sequence Features Regulate the Formation of FG Network at the Center of the Nuclear Pore Complex Peyro M, Soheilypour M, Lee B, Mofrad MRK. Scientific Reports. Vol:5, Article Number:15795, 2015 |
 |
Continuous Distributed Representation of Biological Sequences for Deep Proteomics and Genomics Asgari E, Mofrad MRK. PLoS ONE 10(11): e0141287. doi:10.1371/journal.pone.0141287. |
 |
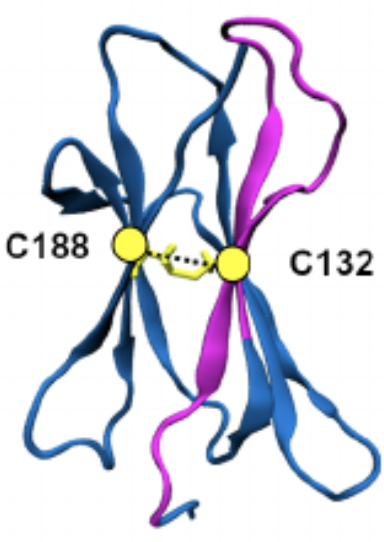
A disulfide bond is required for the transmission of forces through SUN-KASH complexes Jahed Z, Shams H, Mofrad MRK. Biophysical Journal. 2015 Aug 4;109(3):501-509. |
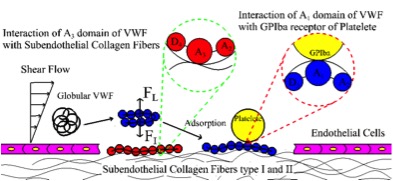 |
Cooperation within Von Willebrand Factors Enhances Adsorption Mechanism Heidari M, Mehrbod M, Ejtehadi MR, Mofrad MRK. Journal of The Royal Society Interface, 12(109), 20150334. |
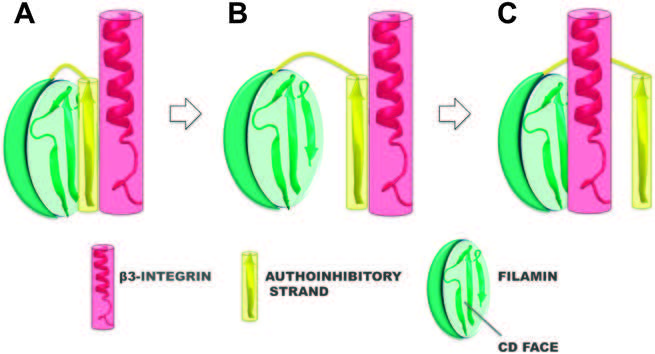 |
Mechanisms of integrin and filamin binding and their interplay with talin during early focal adhesion formation Truong T*, Shams H*, Mofrad MRK. Integrative Biology. 2015 (*co-primary authors) |
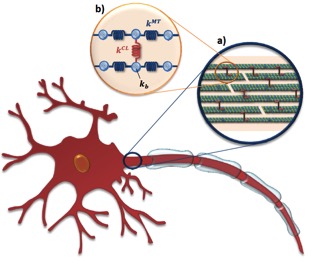 |
Torsional Behavior of Axonal Microtubule Bundles Lazarus C, Soheilypour M, Mofrad MRK. Biophysical Journal, Volume 109, Issue 2, p231–239, 21 July 2015 |
 |
Adhesion characteristics of Staphylococcus aureus bacterial cells on funnel-shaped palladium–cobalt alloy nanostructures cells Gu J, Chen PZ, Seo BB, Jardin JM, Verma MS, Jahed Z, Mofrad MRK, Gu FX, Tsui TY, Journal of Experimental Nanoscience. 2015. |
 |
Trapping polystyrene and latex nanospheres inside hollow nanostructures using Staphylococcus aureus cells Seo BB, Chen PZ, Jahed Z, Mofrad MRK, Gu FX, Tsui TY. Journal of Experimental Nanoscience. (ahead-of-print), 1-11, 2015. |
 |
Buckling Behavior of Individual and Bundled Microtubules Soheilypour M, Peyro M, Peter S, Mofrad MRK. Biophysical Journal, Volume 108, Issue 7, p. 1718–1726, 7 April 2015. |
 |
Directional migration and differentiation of neural stem cells within three-dimensional microenvironments Shamloo A, Heibatollahi M, Mofrad MRK. Integrative Biology, 2015, 7(3), 335-344. |
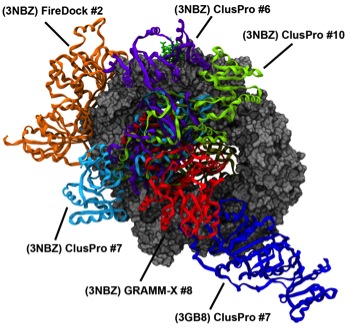 |
The interaction of RNA helicase DDX3 with HIV-1 Rev-CRM1-RanGTP complex during the HIV replication cycle Mahboobi SH, Javanpour AA, Mofrad MRK. PloS one, 2015, 10(2), e0112969. |
 |
An agent-based model for mRNA export through the nuclear pore complex Azimi M*, Bulat E*, Weis K, Mofrad MRK. Molecular Biology of the Cell (MBoC): Special Issue on Quantitative Biology, 2014 Nov 5;25(22):3643-53. |
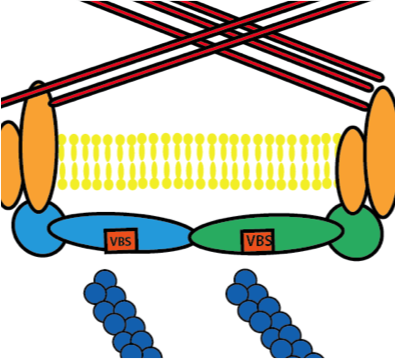 |
The Talin Dimer Structure Orientation Is Mechanically Regulated Golji J, Mofrad MRK. Biophysical Journal, 2014, 107(8), 1802-1809. |
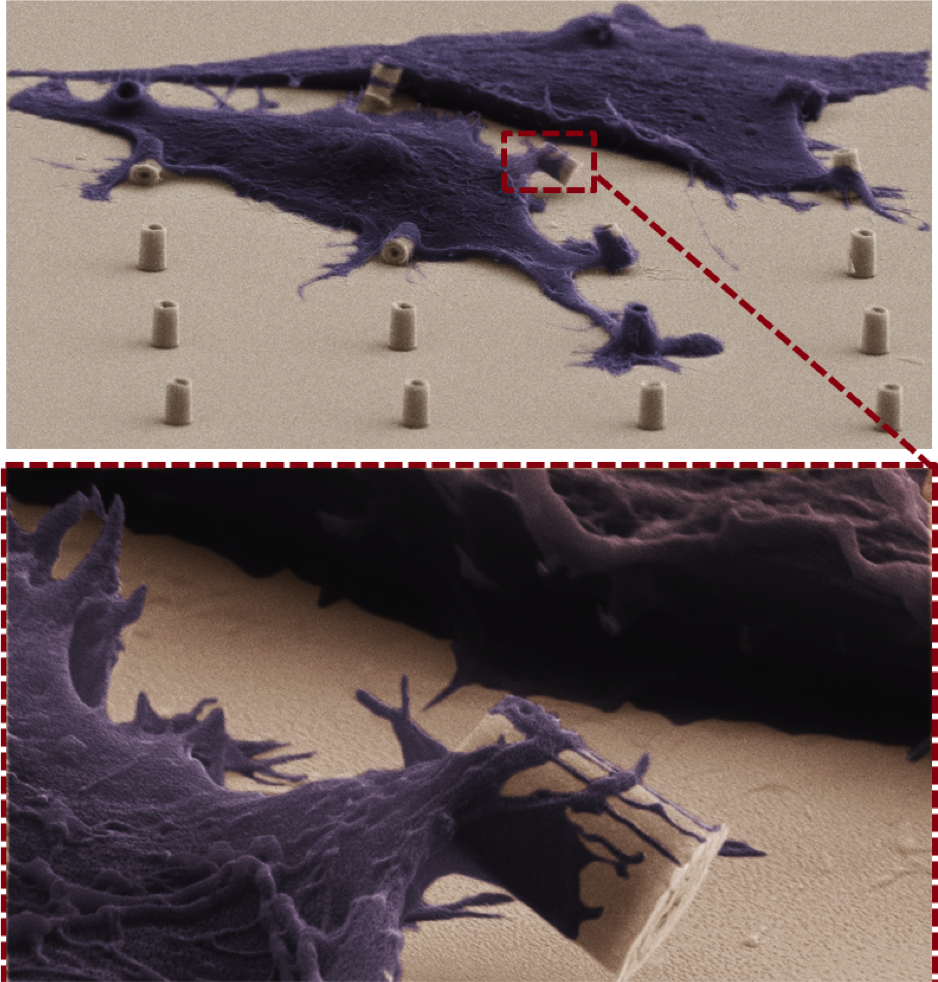 |
Cell responses to metallic nanostructure arrays with complex geometries Jahed Z, Molladavoodi S, Seo BB, Gorbet M, Tsui TY#, Mofrad MRK#. Biomaterials, 2014 Nov;35(34):9363-71. |
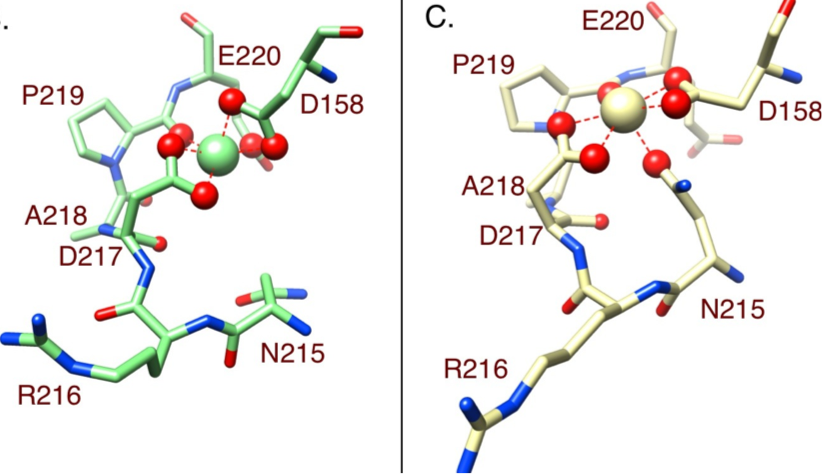 |
The α-subunit regulates stability of the metal ion at the ligand-associated metal-ion binding site in β3 integrins Rui X*, Mehrbod M*, Van Agthoven JF, Xiong JP, Mofrad MRK#, Arnaout MA#. Journal of Biological Chemistry, 2014 Jun 28. [Epub ahead of print]. |
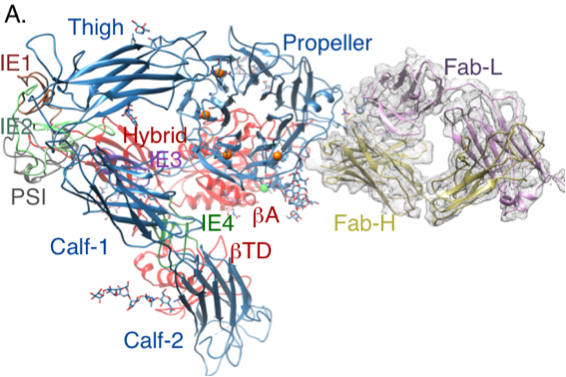 |
Atomic basis for the species-specific inhibition of αV integrins by mAb 17E6 is revealed by the crystal structure of αVβ3 ectodomain-17E6 Fab complex Mahalingam B, Van Agthoven JF, Xiong JP, Alonso JL, Adair BD, Rui X, Anand S, Mehrbod M, Mofrad MRK, Burger D, Goodman SL, Arnaout MA. Journal of Biological Chemistry, 2014 Apr 1. [Epub ahead of print] |
 |
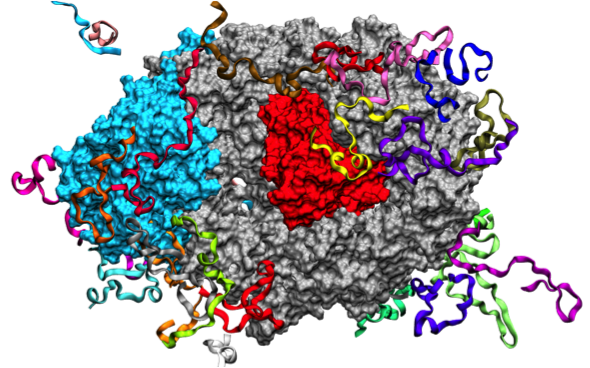
Mechanotransduction Pathways Linking the Extracellular Matrix to the Nucleus Jahed Z, Shams H, Mehrbod M, Mofrad MRK. International Review of Cell and Molecular Biology, 2014;310:171-220. |
 |
The Interaction of CRM1 and the Nuclear Pore Protein Tpr Zhao C*, Mahboobi SH*, Moussavi-Baygi R, Mofrad MRK. PLoS One, April 2014, Volume 9 | Issue 4 | e93709. |
 |
Responses of Staphylococcus Aureus Bacterial Cells to Nanocrystalline Nickel Nanostructures Jahed Z, Lin P, Seo BB, Verma MS, Gu FX, Tsui TY#, Mofrad MRK#. Biomaterials. 2014 Feb 24. doi: 10.1016/j.biomaterials.2014.01.080. |
 |
Filamin: A structural and functional biomolecule with important roles in cell biology, signaling and mechanics Modarres HP, Mofrad MRK. Molecular and Cellular Biomechanics, 2014, 11, 039-065. |
 |
Actin Reorganization through Dynamic Interactions with Single-Wall Carbon Nanotubes Shams H, Holt B, Mahboobi SH, Jahed Z, Islam M, Dahl KN, Mofrad MRK. ACS nano, 2013, 8(1), 188-197. |
 |
Higher Nucleoporin-Importinβ Affinity at the Nuclear Basket Increases Nucleocytoplasmic Import. Azimi M, Mofrad MRK. PLoS One. 2013 Nov 25;8(11):e81741. doi: 10.1371/journal.pone.0081741. |
 |
Quantifying intracellular protein binding thermodynamics during mechanotransduction based on FRET spectroscopy. Abdul Rahim NA, Pelet S, Mofrad MRK, So PTC, Kamm RD. Methods. 2013 Oct 31. doi:pii: S1046-2023(13)00403-9. 10.1016/j.ymeth.2013.10.007. |
 |
Emerging trends in heart valve engineering: part I. Solutions for future. Kheradvar A, Groves EM, Dasi LP, Alavi SH, Tranquillo R, Grande-Allen KJ, Simmons CA, Griffith B, Falahatpisheh A, Goergen CJ, Mofrad MR, Baaijens F, Little SH, Canic S. Annals of Biomedical Engineering, 2015 Apr;43(4):833-43. |
 |
Emerging Trends in Heart Valve Engineering: Part II. Novel and Standard Technologies for Aortic Valve Replacement. Kheradvar A, Groves EM, Goergen CJ, Alavi SH, Tranquillo R, Simmons CA, Dasi LP, Grande-Allen KJ, Mofrad MR, Falahatpisheh A, Griffith B, Baaijens F, Little SH, Canic S. Annals of Biomedical Engineering, 2015 Apr;43(4):844-57. |
 |
Emerging Trends in Heart Valve Engineering: Part III. Novel Technologies for Mitral Valve Repair and Replacement. Kheradvar A, Groves EM, Simmons CA, Griffith B, Alavi SH, Tranquillo R, Dasi LP, Falahatpisheh A, Grande-Allen KJ, Goergen CJ, Mofrad MR, Baaijens F, Canic S, Little SH. Annals of Biomedical Engineering, 2015 Apr;43(4):858-70. |
 |
Emerging Trends in Heart Valve Engineering: Part IV. Computational Modeling and Experimental Studies Kheradvar A, Groves EM, Falahatpisheh A, Mofrad MRK, Alavi SH, Tranquillo R, Dasi LP, Simmons CA, Grande-Allen KJ, Goergen CJ, Baaijens F, Little SH, Canic S, Griffith B. Annals of Biomedical Engineering, 2015. |
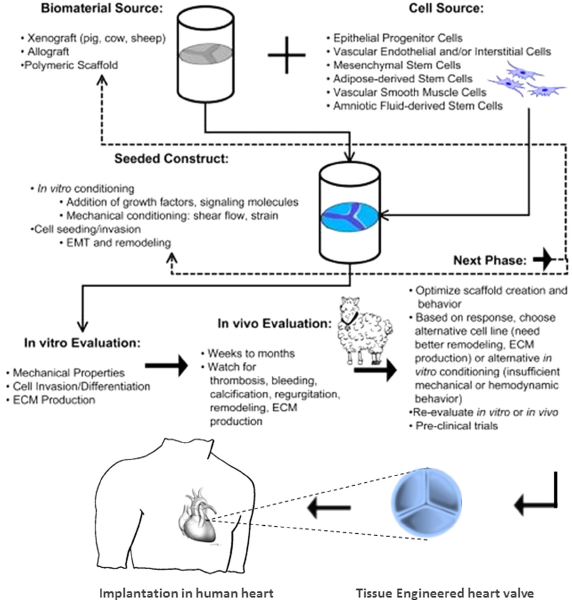 |
Biomechanical properties of native and tissue engineered heart valve constructs. Hasan A, Ragaert K, Swieszkowski W, Selimović S, Paul A, Camci-Unal G, Mofrad MRK, Khademhosseini A. Journal of Biomechanics. 2013 Oct 21. doi:pii: S0021-9290(13)00446-6. 10.1016/j.jbiomech.2013.09.023. |
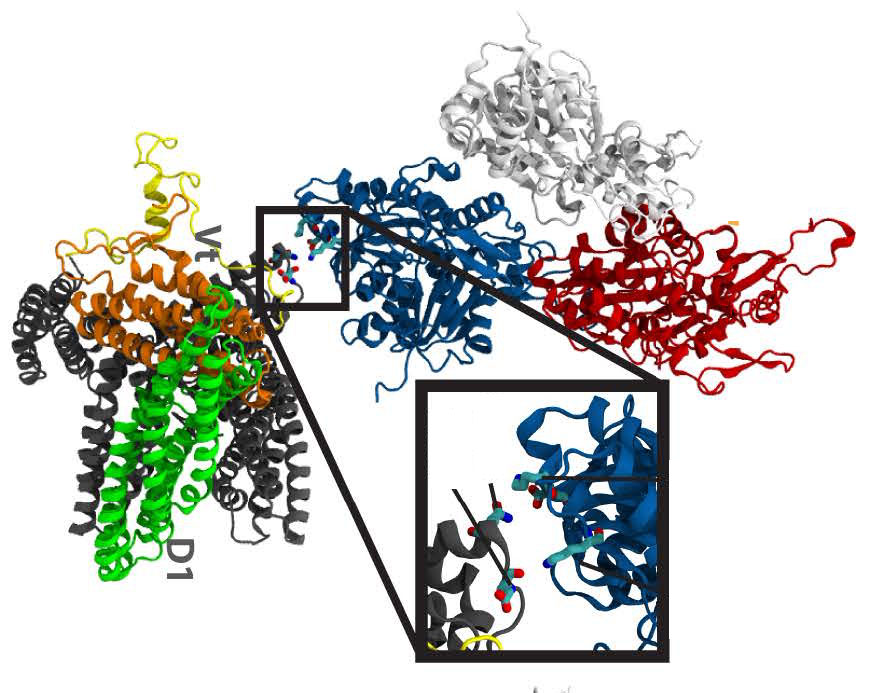
 |
On the Activation of Integrin αIIbβ3: Outside-In and Inside-Out Pathways. Mehrbod M, Trisno S, Mofrad MRK. Biophysical Journal, 2013 Sept; 105(6). |
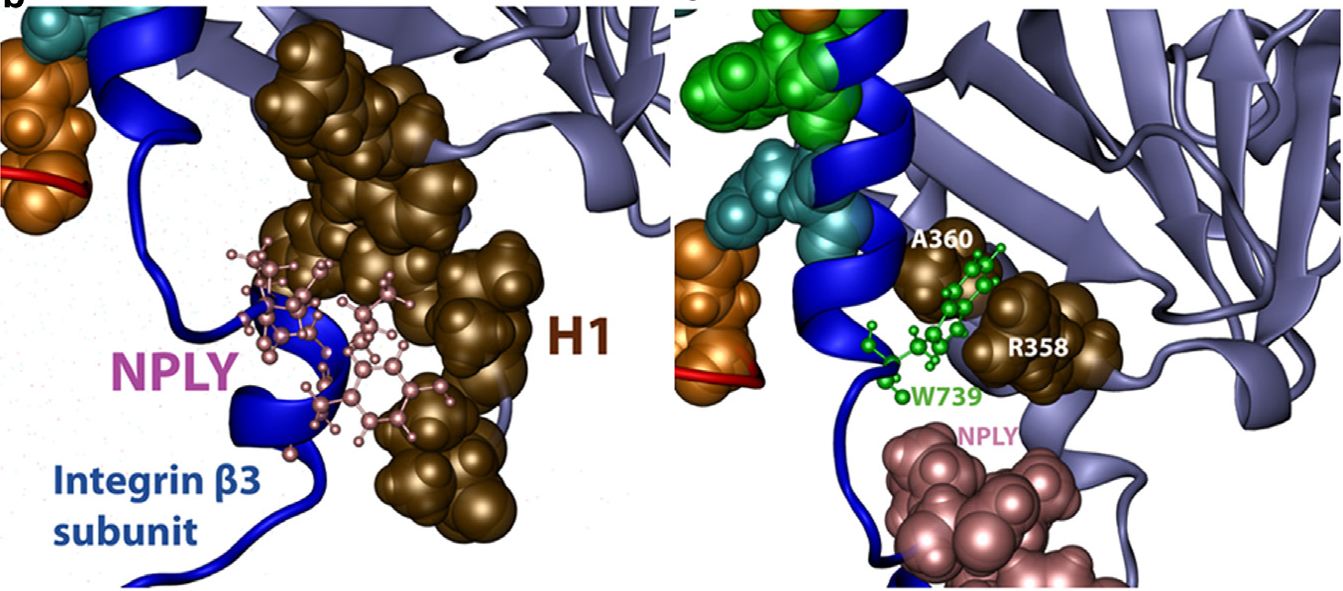
 |
An Agent Based Model of Integrin Clustering: Exploring the Role of Ligand Clustering, Integrin Homo-Oligomerization, Integrin-Ligand Affinity, Membrane Crowdedness and Ligand Mobility. Jamali Y, Jamali T, Mofrad MRK. Journal of Computational Physics. 2013; 244:264–278 |
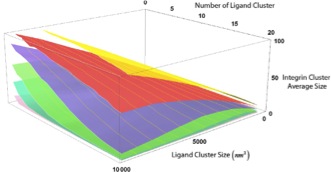
 |
The Interaction of Vinculin with Actin. Golji J, Mofrad MRK. PLoS Computational Biology, 2013 Apr;9(4):e1002995. |
 |
Localized Lipid Packing of Transmembrane Domains Impedes Integrin Clustering. Mehrbod M, Mofrad MRK. PLoS Computational Biology, 2013 Mar; 9(3): e1002948. |
 |
Molecular Trajectory of Alpha-Actinin Activation. Shams H, Golji J, Mofrad MRK. Biophysical Journal, November 2012, 103(10):2050-2059. |
 |
Altered Cell Mechanics from the Inside: Dispersed Single Wall Carbon Nanotubes Integrate with and Restructure Actin. Holt BD, Shams H, Horst TA, Basu S, Rape DA, Wang Y-L, Rohde GK, Mofrad MRK, Islam MF and Dahl KN. Journal of Functional Biomaterials, 2012 May; 3:398-417. |
 |
Phosphorylation Primes Vinculin for Activation. Golji J, Wendorff TJ, Mofrad MRK. Biophysical Journal, 2012 May; 102:2022-2030. |
 |
Passive control of cell locomotion using micropatterns: the effect of micropattern geometry on the migratory behavior of adherent cells. Lab Chip. Yoon SH, Kim YK, Han ED, Seo YH, Kim BH, Mofrad MRK. 2012 Jul 7;12(13):2391-402[highlighted in Lab Chip Research Highlights, 2012 Jul 7;12(13):2391-4023.] |
 |
Computational Modeling of Axonal Microtubule Bundles under Tension. Peter SJ, Mofrad MRK. Biophysical Journal, 2012 February; 102(3):749-757. |
 |
Biophysical Coarse-Grained Modeling Provides Insights into Transport through the Nuclear Pore Complex. Moussavi-Baygi RM, Jamali Y, Karimi R, Mofrad MRK. Biophysical Journal. 2011; 100(6):1410-1419[cover]. |
 |
Brownian Dynamics Simulation of Nucleocytoplasmic Transport: A Coarse-Grained Model for the Functional State of the Nuclear Pore Complex. Moussavi-Baygi RM, Jamali Y, Karimi R, Mofrad MRK. PLoS Computational Biology. 2011 June; 7(6): e1002049. |
 |
Nuclear Pore Complex: Biochemistry and Biophysics of Nucleocytoplasmic Transport in Health and Disease. Jamali T, Jamali Y, Mehrbod M, Mofrad MRK. International Review of Cell and Molecular Biology, 2011; 287: 233-286. |
 |
On the Significance of Microtubule Flexural Behavior in Cytoskeletal Mechanics. Mehrbod M, Mofrad MRK. PLoS One. 2011; 6(10):e25627. |
 |
Accounting for Diffusion in Agent Based Models of Reaction-Diffusion Systems with Application to Cytoskeletal Diffusion. Azimi M, Jamali Y, Mofrad MRK. PLoS One. 2011;6(9):e25306. |
 |
A biological breadboard platform for cell adhesion and detachment studies. Yoon SH, Chang J, Lin L, Mofrad MRK. Lab on a Chip, 2011; 11(20):3555-62[highlighted in Lab Chip Research Highlights, December 21, 2011, 11(24):4141-3]. |
 |
Cell adhesion and detachment on gold surfaces modified with a thiol-functionalized RGD peptide. Yoon SH, Mofrad MRK. Biomaterials, 2011; 32: 7286-7296. |
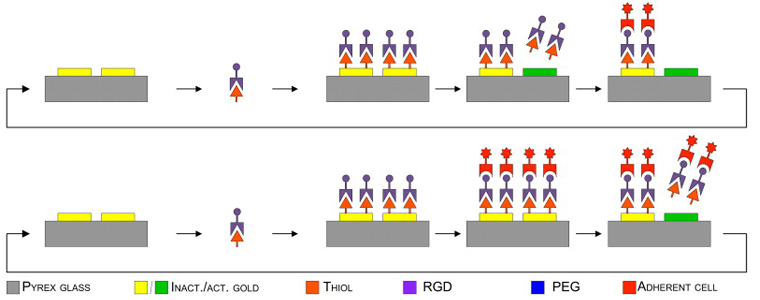 |
Viscoelastic characterization of the retracting cytoskeleton using subcellular detachment. Yoon SH, Lee C, Mofrad MRK. Applied Physics Letters, 2011; 98, 133701-3. |
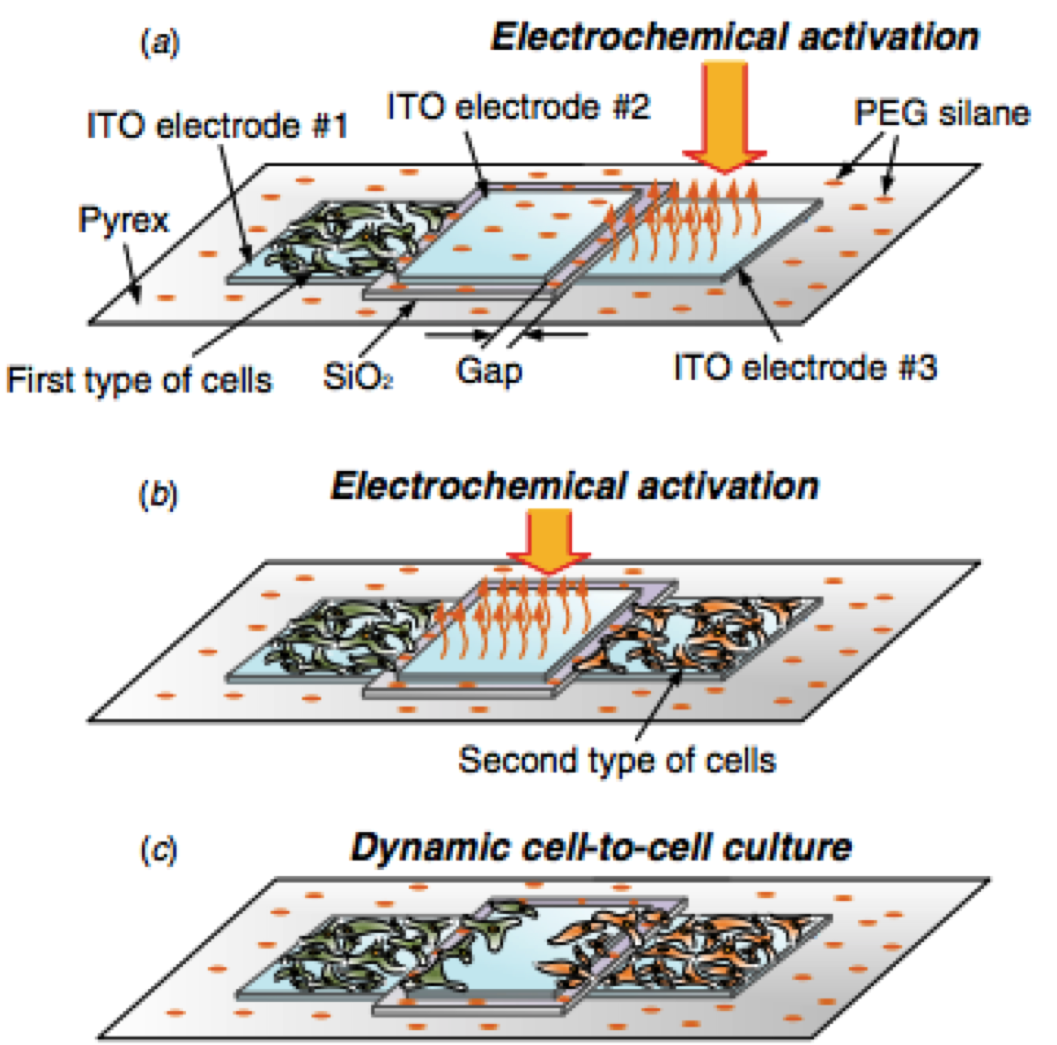 |
MEMS Based Dynamic Cell-to-Cell Culture Platforms Using Electrochemical Surface Modifications. Chang J, Yoon SH, Mofrad MRK, Lin L. Journal of Micromechanics and Microengineering, 2011; 21, 054028. |
 |
Vinculin Activation Is Necessary for Complete Talin Binding. Golji J, Lam J, Mofrad MRK. Biophysical Journal 2011 Jan 19;100(2):332-40. |
 |
A Sub-Cellular Viscoelastic Model for Cell Population Mechanics. Jamali Y, Azimi, M, Mofrad MRK. PLoS One. 2010; 5(8):e12097 |
 |
Analysis of Circular PDMS Microballoons with Hyper-Deflection for MEMS Design. Yoon S, Reyes-Ortiz V, Kim K, Seo YH, Mofrad MRK. J. MEMS. 2010. 19(4):854-864. |
 |
On the multiscale modeling of heart valve biomechanics in health and disease. Weinberg EJ, Shahmirzadi D, Mofrad MR. Biomech Model Mechanobiol. 2010 Aug;9(4):373-87. |
 |
Liver-assist device with a microfluidics-based vascular bed in an animal model. Hsu WM, Carraro A, Kulig KM, Miller ML, Kaazempur-Mofrad M, Weinberg E, Entabi F, Albadawi H, Watkins MT, Borenstein JT, Vacanti JP, Neville C. Ann Surg. 2010 Aug;252(2):351-7. |
 |
Pulmonary tissue engineering using dual-compartment polymer scaffolds with integrated vascular tree. Fritsche CS, Simsch O, Weinberg EJ, Orrick B, Stamm C, Kaazempur-Mofrad MR, Borenstein JT, Hetzer R, Vacanti JP. Int J Artif Organs. 2009 Oct;32(10):701-10. |
 |
A Molecular Dynamics Investigation of Vinculin Activation. Golji J, Mofrad MRK. Biophysical Journal. 2010 August; 99(3): 1073–1081. |
 |
Averaged Implicit Hydrodynamic Model of Semiflexible Filaments. Chandran PL, Mofrad MRK. Physical Rev. E 2010 Mar;81(3 Pt 1):031920. |
 |
Molecular Biomechanics: The Molecular Basis of How Forces Regulate Cellular Function. Bao G, Kamm RD, Thomas W, Hwang W, Fletcher DA, Grodzinsky A, Zhu C, Mofrad MRK. Cellular & Molecular Bioeng. 2010; 3(2):91-105. |
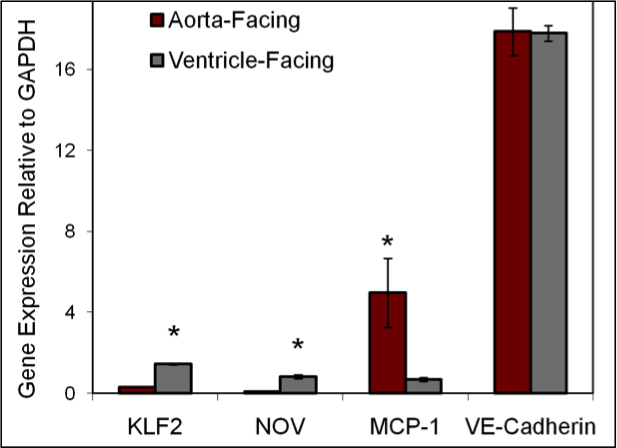 |
Hemodynamic environments from opposing sides of human aortic valve leaflets evoke distinct endothelial phenotypes in vitro. Weinberg EJ, Mack PJ, Schoen FJ, Garcia-Cardena G, Kaazempur Mofrad MR. Cardiovasc Eng. 2010 Mar;10(1):5-11. |
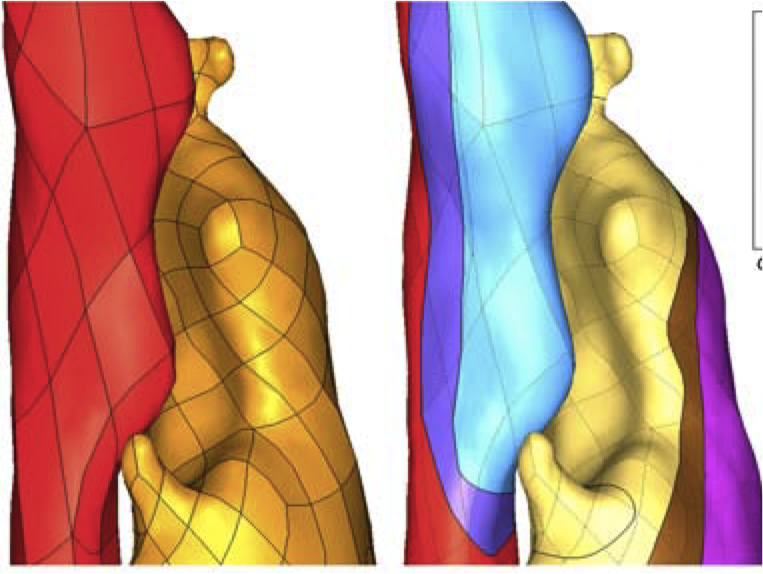 |
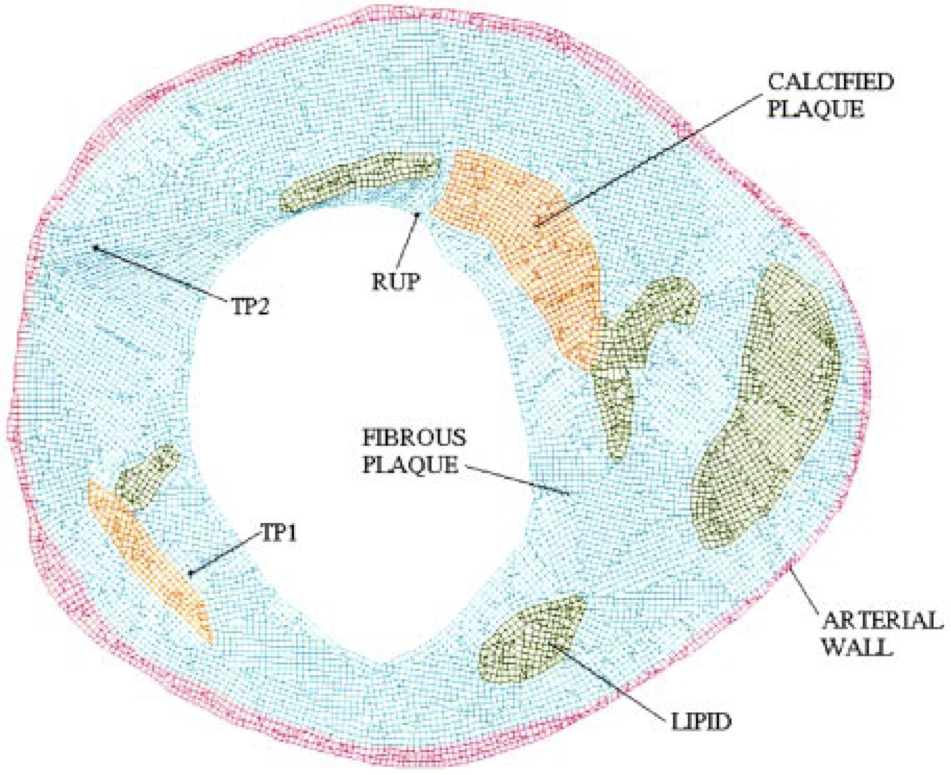
Carotid atheroma rupture observed in vivo and FSI-predicted stress distribution based on pre-rupture imaging. Leach JR, Rayz VL, Soares B, Wintermark M, Mofrad MR, Saloner D. Ann Biomed Eng. 2010 Aug;38(8):2748-65. |
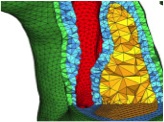 |
An efficient two-stage approach for image-based FSI analysis of atherosclerotic arteries. Leach JR, Rayz VL, Mofrad MR, Saloner D. Biomech Model Mechanobiol. 2010 Apr;9(2):213-23. |
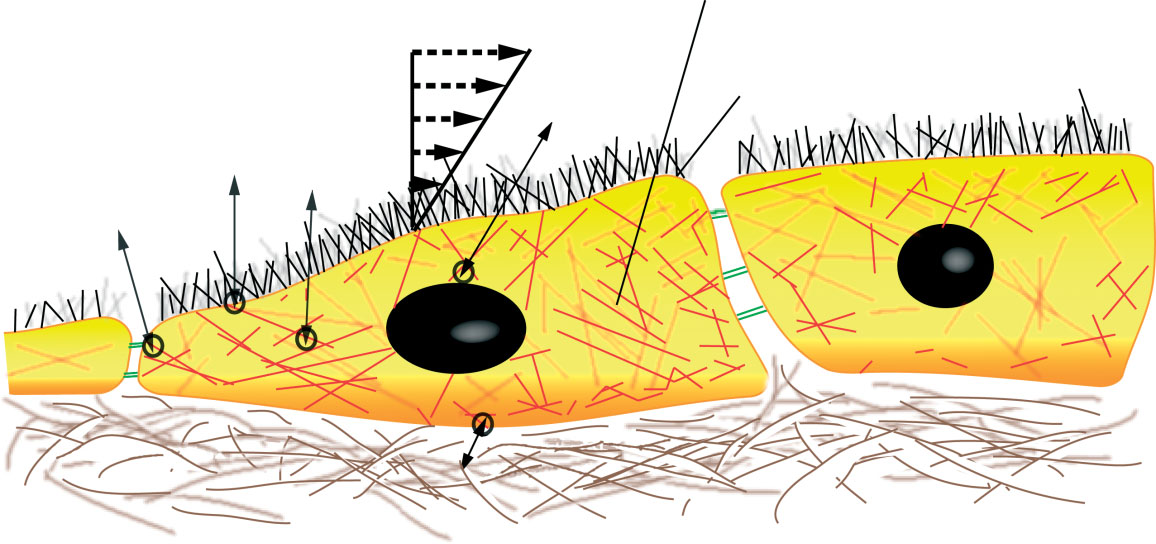 |
Mechanotransduction: a major regulator of homeostasis and development. Kolahi KS, Mofrad MR. Wiley Interdiscip Rev Syst Biol Med. 2010 Nov-Dec;2(6):625-39. |
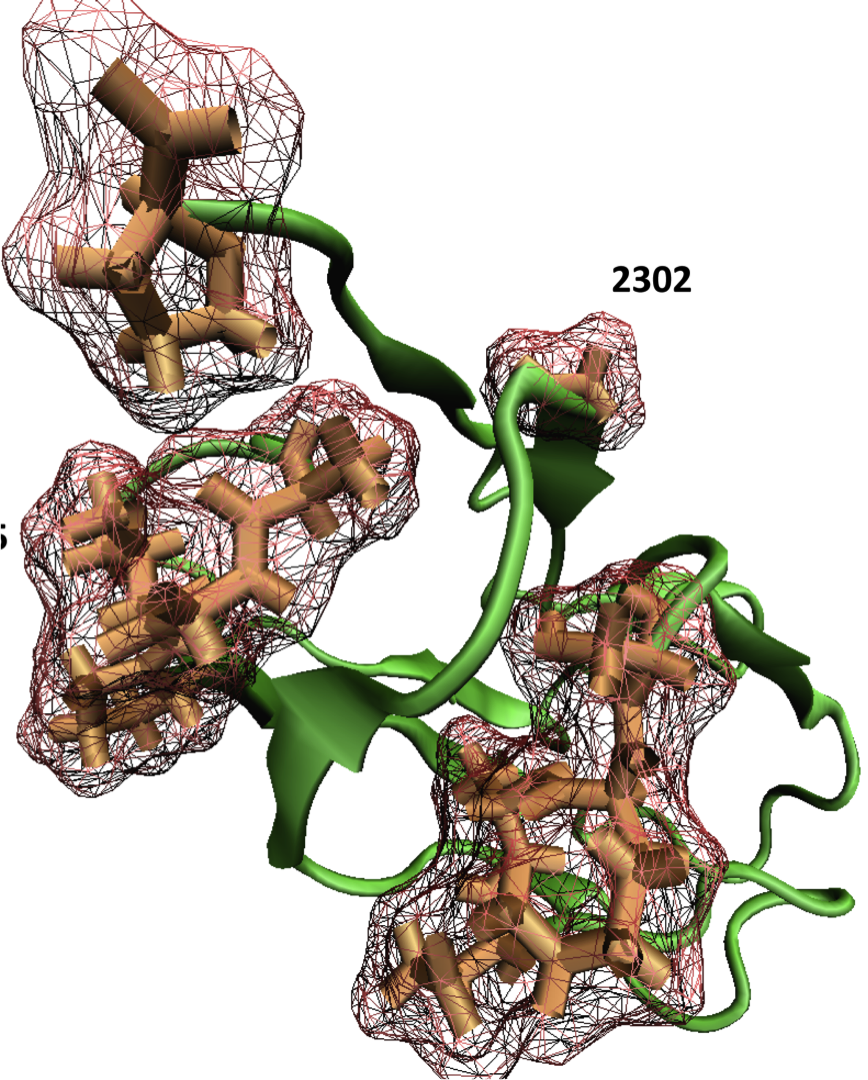 |
Phosphorylation Facilitates the Integrin Binding of Filamin Under Force. Chen HS, Kolahi KS, Mofrad MRK. Biophysical Journal 2009 Dec 16;97(12):3095-104. |
 |
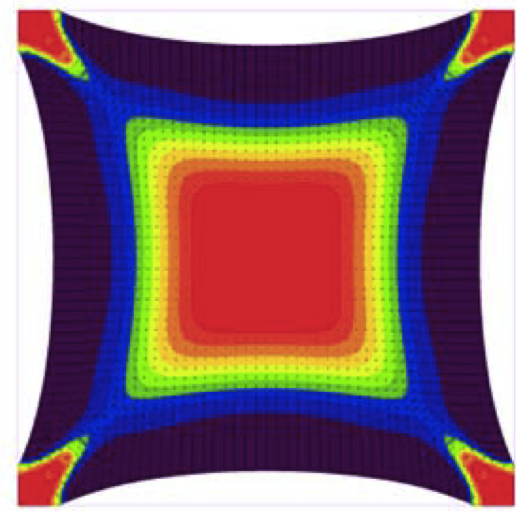
Rheology of the cytoskeleton. Mofrad MRK. Ann. Rev. of Fluid Mechanics. 2009; 41:433-453. |
 |
Band-like stress fiber propagation in a continuum and implications for myosin contractile stresses. Chandran PL, Wolf CB, Mofrad MRK. 2009; 2(1):13-27. |
 |
A computational model of aging and calcification in the aortic heart valve. Weinberg EJ, Schoen FJ, Mofrad MR. PLoS One. 2009 Jun 18;4(6):e5960. |
 |
Molecular mechanics of the alpha-actinin rod domain: bending, torsional, and extensional behavior. Golji J, Collins R, Mofrad MR. PLoS Comput Biol. 2009 May;5(5):e1000389. |
 |
Quantitative analysis of epithelial morphogenesis in Drosophila oogenesis: New insights based on morphometric analysis and mechanical modeling. Kolahi KS, White PF, Shreter DM, Classen AK, Bilder D, Mofrad MR. Dev Biol. 2009 Jul 15;331(2):129-39. |
 |
Rods-on-string idealization captures semiflexible filament dynamics. Chandran PL, Mofrad MR. Phys Rev E Stat Nonlin Soft Matter Phys. 2009 Jan;79(1 Pt 1):011906. |
 |
Estimation of nonlinear mechanical properties of vascular tissues via elastography. Karimi R, Zhu T, Bouma BE, Mofrad MR. Cardiovasc Eng. 2008 Dec;8(4):191-202. |
 |
A multiscale computational comparison of the bicuspid and tricuspid aortic valves in relation to calcific aortic stenosis. Weinberg EJ, Kaazempur Mofrad MR. J Biomech. 2008 Dec 5;41(16):3482-7. |
 |
Concept and computational design for a bioartificial nephron-on-a-chip. Weinberg E, Kaazempur-Mofrad M, Borenstein J. Int. J. Artificial Organs. 2008 Jun;31(6):508-14. |
 |
In vitro analysis of a hepatic device with intrinsic microvascular-based channels. Carraro A, Hsu WM, Kulig KM, Cheung WS, Miller ML, Weinberg EJ, Swart EF, Kaazempur-Mofrad M, Borenstein JT, Vacanti JP, Neville C. Biomed Microdevices. 2008 Dec;10(6):795-805. |
 |
On the octagonal structure of the nuclear pore complex: insights from coarse-grained models. Wolf C, Mofrad MR. Biophys J. 2008 Aug;95(4):2073-85.[cover]. |
 |
Molecular dynamics study of talin-vinculin binding. Lee SE, Chunsrivirot S, Kamm RD, Mofrad MR. Biophys J. 2008 Aug;95(4):2027-36. |
 |
On the cytoskeleton and soft glassy rheology. Mandadapu KK, Govindjee S, Mofrad MR. J Biomech. 2008;41(7):1467-78. |
 |
Transient, three-dimensional, multiscale simulations of the human aortic valve. Weinberg EJ, Kaazempur Mofrad MR. Cardiovasc Eng. 2007 Dec;7(4):140-155. |
 |
Molecular mechanics of filamin’s rod domain. Kolahi KS, Mofrad MR. Biophys J. 2008 Feb 1;94(3):1075-83. |
 |
Microfluidic environment for high density hepatocyte culture. Zhang MY, Lee PJ, Hung PJ, Johnson T, Lee LP, Mofrad MR. Biomedical Microdevices. 2008 Feb;10(1):117-21. |
 |
Microfabrication of three-dimensional engineered scaffolds. Borenstein JT, Weinberg EJ, Orrick BK, Sundback C, Kaazempur-Mofrad MR, Vacanti JP. Tissue Eng. 200LE Aug;13(8):1837-44. |
 |
Force-induced activation of Talin and its possible role in focal adhesion mechanotransduction. Lee SE, Kamm RD, Mofrad MRK. Journal of Biomechanics, 2007;40(9):2096-106. |
 |
Mechanics and Deformation of the Nucleus in Micropipette Aspiration Experiment. Vaziri A, Kaazempur Mofrad MR. Journal of Biomechanics, 2007;40(9):2053-62. |
 |
A Computational Study on Power-Law Rheology of Soft Glassy Materials with Application to Cell Mechanics Vaziri A, Xue Z, Kamm RD, Kaazempur Mofrad MR. Computer Methods in Applied Mechanics and Engineering, 196: 2965-2971, 2007. |
 |
A finite shell element for heart mitral valve leaflet mechanics, with large deformations and 3D constitutive material model Weinberg EJ, Kaazempur Mofrad MR. Journal of Biomechanics, 2007;40(3):705-11. |
 |
A Combined FEM/Genetic Algorithm for Vascular Soft Tissue Elasticity Estimation Khalil AS, Bouma BE, Kaazempur Mofrad MR. Cardiovasc Eng. 2006 Sep;6(3):93-102. |
 |
Deformation of the cell Nucleus under Indentation: Mechanics and Mechanisms Vaziri A, Lee H, Kaazempur Mofrad MR. Journal of Material Research, 2006 Aug; 21(8):1-10. |
 |
A Coarse-grained Model for Force-induced Protein Deformation and Kinetics Karcher H, Lee SE, Kaazempur-Mofrad MR, Kamm RD. Biophysical Journal, 2006 Apr 15;90(8):2686-97. |
 |
A Large-Strain Finite Element Formulation for Biological Tissues with Application to Mitral Valve Leaflet Tissue Mechanics Weinberg EJ, Kaazempur-Mofrad MR. Journal of Biomechanics, 2006;39(8):1557-61.. |
 |
Non-rigid registration for fusion of carotid vascular ultrasound and MRI volumetric datasets Chan RC, Sokka S, Hinton D, Houser S, Manzke R, Hanekamp A, Reddy VY, Kaazempur-Mofrad MR, and Rasche V. Proc. SPIE Vol. 6144 61442E1-8. |
 |
Tissue Elasticity Estimation with Optical Coherence Elastography: Toward Mechanical Characterization of In Vivo Soft Tissue Khalil AS, Chan RC, Chau AH, Bouma BE, Kaazempur-Mofrad MR. Annals of Biomedical Engineering, 2005 Nov;33(11):1631-9.. |
 |
Mass Transport and Fluid Flow in Stenotic Arteries: Axisymmetric and Asymmetric Models Kaazempur-Mofrad MR, Wada S, Myers JG, and Ethier CR. International Journal of Heat and Mass Transfer, 2005; 48:4510-4517. |
 |
Exploring the molecular basis for mechanosensation, signal transduction, and cytoskeletal remodeling Kaazempur-Mofrad MR, Abdul-Rahim NA, Karcher H, Mack PJ, Yap B, Kamm RD. Acta BioMaterial, 2005 May;1(3):281-93. |
 |
Endothelialized Microvasculature Based on a Biodegradable Elastomer Fidkowski C, Kaazempur-Mofrad MR, Borenstein JT, Vacanti JP, Langer R, Wang Y. Tissue Engineering, Engineering, 2005 Jan-Feb;11(1-2):302-9. |
 |
On the Constitutive Models for Heart Valve Leaflet Mechanics. Weinberg EJ, Kaazempur-Mofrad MR. Cardiovascular Engineering, 2005; 5(1): 37-43. |
 |
Endothelialized networks with a vascular geometry in microfabricated poly(dimethyl siloxane) Shin M, Matsuda K, Ishii O, Terai H, Kaazempur-Mofrad M, Borenstein J, Detmar M, Vacanti JP. Biomedical Microdevices, 2004 Dec;6(4):269-78. |
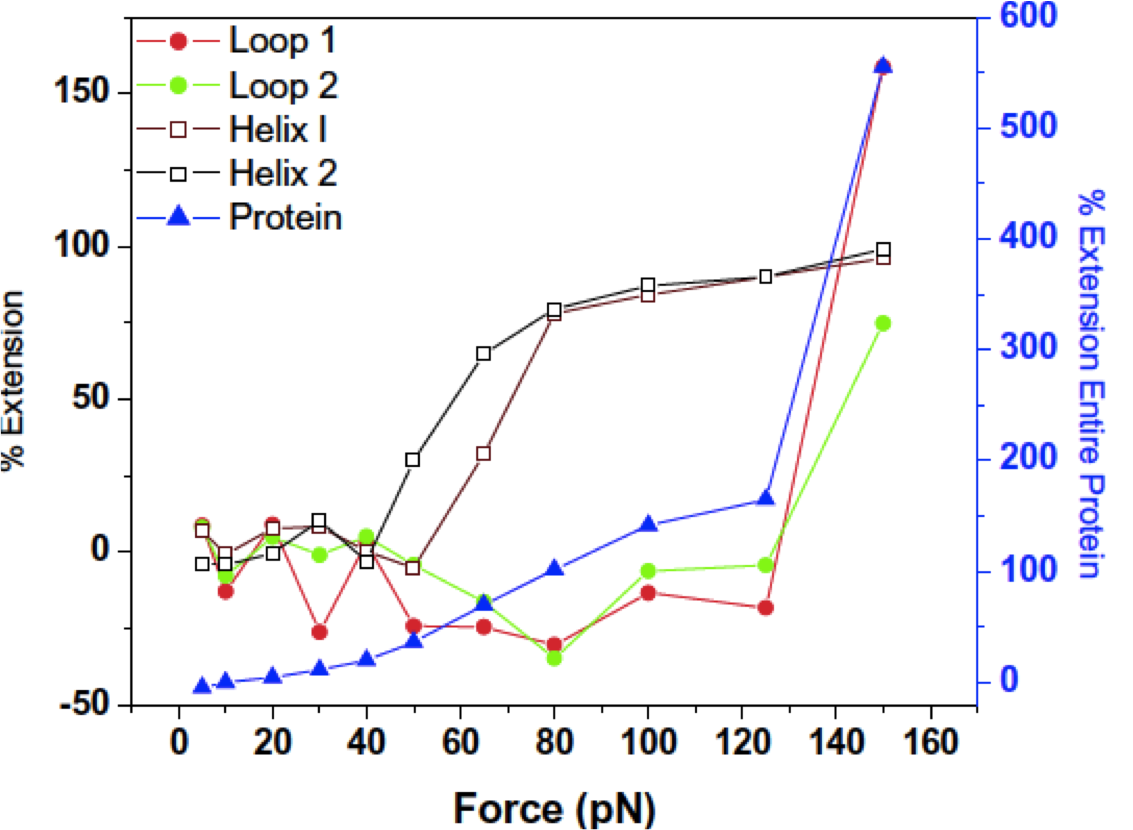 |
How Flexible is α-Actinin’s Rod Domain? Zaman MH, Kaazempur-Mofrad MR. Mechanics & Chemistry of Biosystems. 2004 Dec;1(4):291-302. |
 |
Force-induced Conformational Changes in Focal Adhesion Targeting Region of Focal Adhesion Kinase: A Steered Molecular Dynamics Study Kaazempur-Mofrad MR, Golji J, Abdul Rahim NA, Kamm RD. Mechanics & Chemistry of Biosystems. 2004 Dec;1(4):253-65. |
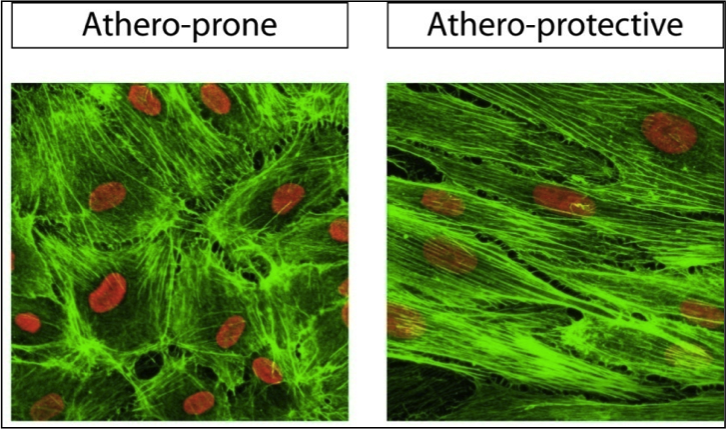 |
Distinct Endothelial Phenotypes Evoked by Arterial Waveforms Derived from Atherosclerosis-Susceptible and Resistant Regions of Human Vasculature Dai G, Kaazempur-Mofrad MR, Natarajan S, Zhang Y, Vaughn S, Blackman BR, Kamm RD, García-Cardena G, Gimbrone MA, Jr. PNAS, 2004 Oct 12;101(41):14871-6. |
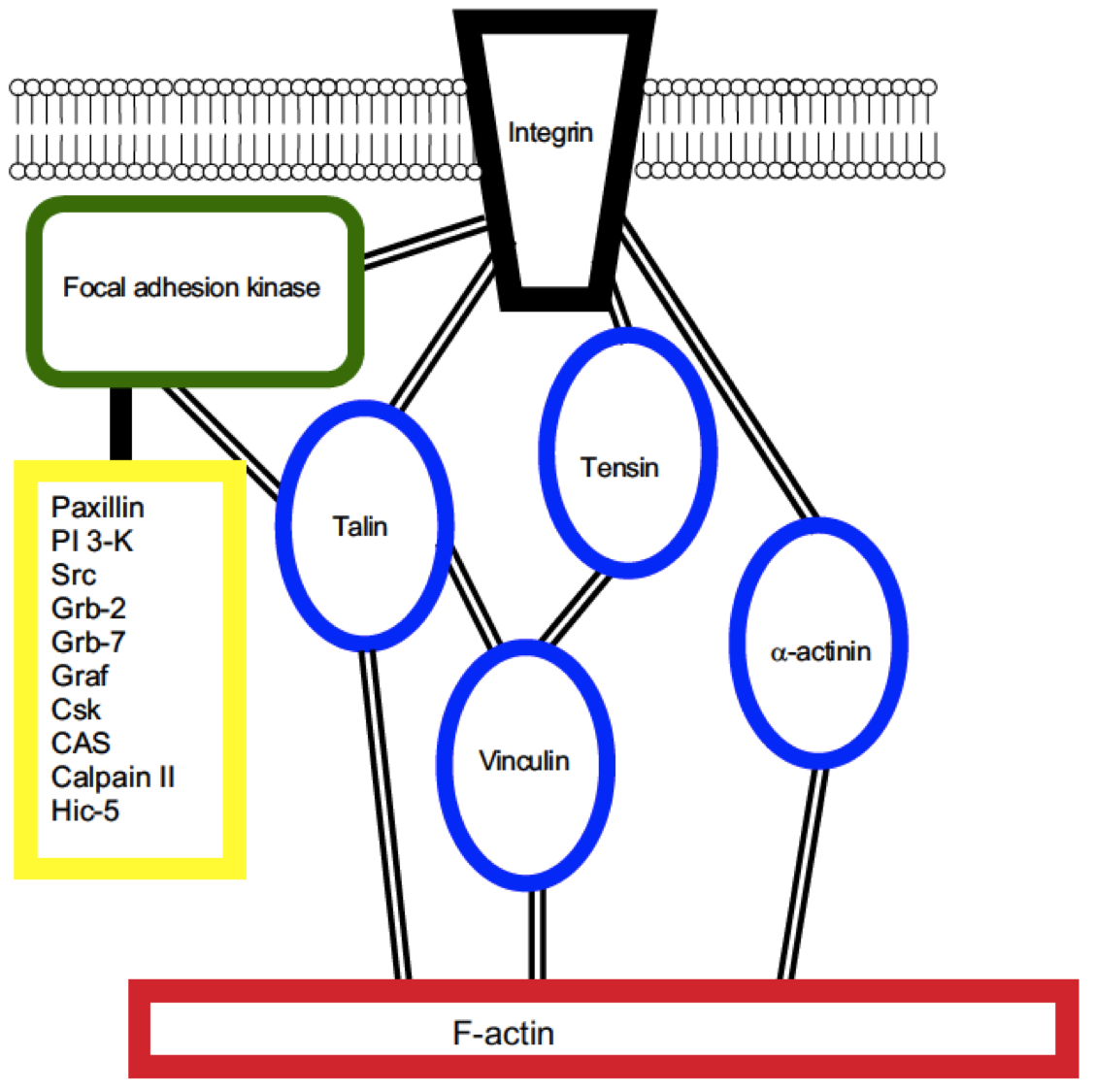 |
On the Molecular Basis for Mechanotransduction Kamm RD and Kaazempur-Mofrad MR. Mechanics & Chemistry of Biosystems, 2004 Sep;1(3):201-209. |
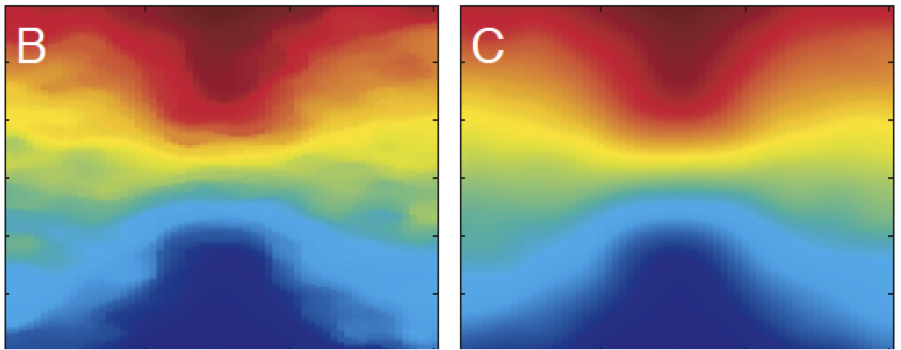 |
OCT-based Arterial Elastography: Robust Estimation exploiting Tissue Biomechanics Chan RC, Chau AH, Karl WC, Nadkarni S, Khalil AS, Shishkov M, Tearney GJ, Kaazempur-Mofrad MR, Bouma BE. Optics Express, 2004 Sep 20;12(19):4558-72. |
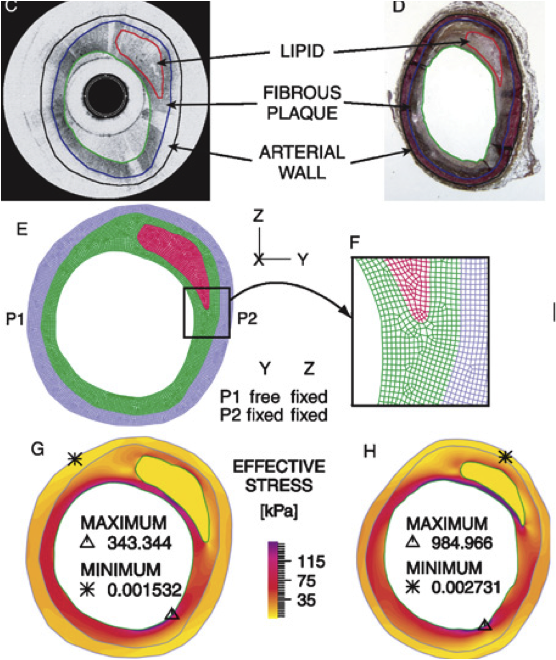 |
Mechanical Analysis of Atherosclerotic Plaques based on Optical Coherence Tomography Chau AH, Chan RC, Shishkov M, MacNeill, B, Iftima N, Tearney GJ, Kamm RD, Bouma B, Kaazempur-Mofrad MR. Annals of Biomedical Engineering, 2004 Nov;32(11):1494-503. |
 |
Biodegradable Microfluidics King KR, Wang CJ, Kaazempur-Mofrad MR, Vacanti JP, Borenstein JT. Advanced Materials, 2004; 16(22):2007-2012. |
 |
Force-induced Focal Adhesion Translocation: Effects of Force Amplitude and Frequency Mack PJ, Kaazempur-Mofrad MR, Karcher H, Lee RT, Kamm RD. American Journal of Physiology – Cell Physiology, 2004 Oct;287(4):C954-62 . |
 |
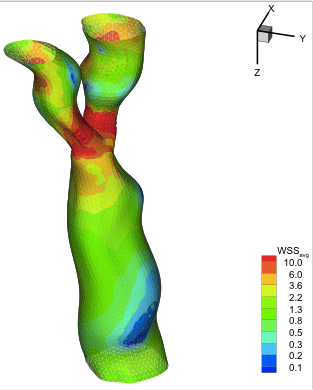
Hemodynamics and wall mechanics in human carotid bifurcation and its consequences for atherogenesis: investigation of inter-individual variation. Younis HF, Kaazempur-Mofrad MR, Chan RC, Isasi AG, Hinton DP, Chau AH, Kim LA, Kamm RD. Biomech Model Mechanobiol. 2004 Sep;3(1):17-32. |
 |
Characterization of the Atherosclerotic Carotid Bifurcation using MRI, Finite Element Modeling and Histology Kaazempur-Mofrad MR, Younis HF, Isasi AG, Chan RC, Hinton DP, Sukhova G, LaMuraglia GM, Lee RT and Kamm RD. Annals of Biomedical Engineering, 2004 July;32(7):932-46. |
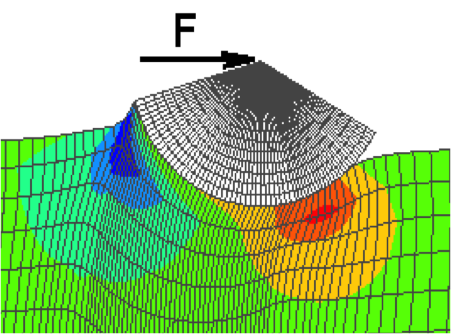 |
A Three-Dimensional Viscoelastic Model for Cell Deformation with Experimental Validation Karcher H, Lammerding J, Huang H, Lee RT, Kamm RD, and Kaazempur-Mofrad MR. Biophysical Journal, 2003 Nov;85(5):3336-49. |
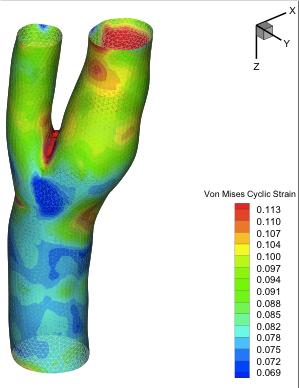 |
Cyclic Strain in Human Carotid Bifurcation and its Potential Correlation to Atherogenesis: Idealized and Anatomically-Realistic Models Kaazempur-Mofrad MR, Younis HF, Patel S, Isasi AG, Chung C, Chan RC, Hinton DP, Lee RT, Kamm RD. Journal of Engineering Mathematics, 2003; 47(3-4):299-314. |
 |
Role of simulation in understanding biological systems. Kaazempur-Mofrad MR, Bathe M, Karcher H, Younis HF, Seong HC, Shim EB, Chan RC, Hinton DP, Powers MJ, Griffith LG, and Kamm RD. Computer & Structures, 2003; 81(8):715-726. |
 |
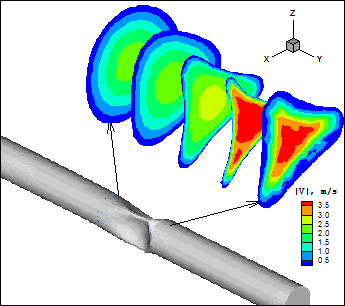
Computational Analysis of the Effects of Exercise on Hemodynamics in the Carotid Bifuration Younis HF, Kaazempur-Mofrad MR, Chan R, Chung C, Kamm RD. Annals of Biomedical Engineering, 2003 Sep;31(8):995-1006. |
 |
A Dynamic Rotational Seeding and Cell Culture System for Vascular Tube Formation. Nasseri BA, Pomeranteseva I, Kaazempur-Mofrad MR, Perry T, Ochoa E, Thomson CA, Oesterle SN, Vacanti JP. Tissue Engineering, 2003 Apr;9(2):291-9. |
 |
On the Sensitivity of Wall Stresses in Diseased Arteries to Variable Material Properties Williamson SD, Lam Y, Younis HF, Huang H, Patel S, Kaazempur-Mofrad MR, Kamm RD. Journal of Biomechanical Engineering, 2003 Feb;125(1):147-55. |
 |
A Characteristic/Finite Element Algorithm for Time-Dependent 3-D Advection-Dominated Transport using Unstructured Grids Kaazempur-Mofrad MR, Minev PD and Ethier CR. Computer Methods in Applied Mechanics and Engineering, 2003; 192(11-12):1281-1298. |
 |
An Efficient Characteristic Galerkin Scheme for the Advection Equation in 3-D. Kaazempur-Mofrad MR and Ethier CR. Computer Methods in Applied Mechanics and Engineering, 2002; 191(46):5345-5363. |
 |
Microfabrication Technology for Vascularized Tissue Engineering. Borenstein JT, Terai H, King KR, Weinberg EJ, Kaazempur-Mofrad MR and Vacanti JP, Biomedical Microdevices, 2002 July; 4(3):167-175. |
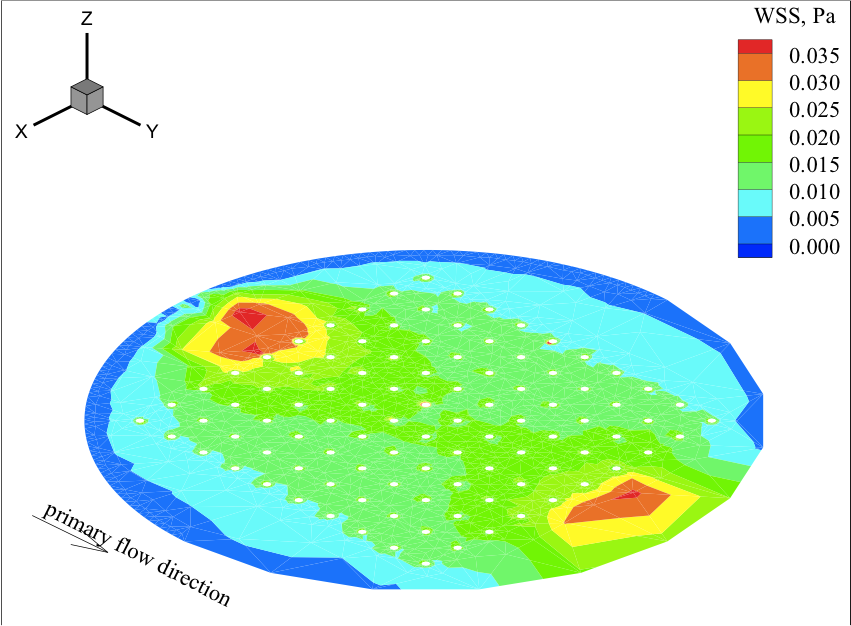 |
A Microfabricated Array Bioreactor for Perfused 3D Liver Culture Powers, MJ, Domansky K, Kaazempur-Mofrad MR, Kalezi A, Capitano A, Upadhyaya A, Kurzawski P, Wack KE, Stolz DB, Kamm RD, Griffith LG. Biotechnology & Bioengineering, 2002 May 5;78(3):257-69. |
 |
Mass Transport in an Anatomically Realistic Human Right Coronary Artery Kaazempur-Mofrad MR and Ethier CR. Annals of Biomedical Engineering, 2001 Feb;29(2):121-7. |
BOOK CHAPTERS (selected)
 |
Tissue Engineering of Microvascular Networks. Borenstein JT, Weinberg EJ, Kaazempur-Mofrad MR, and Vacanti JP. in Encyclopedia of Biomaterials and Biomedical Engineering (Eds.: Wnek GE and Bowlin GL), Marcel Dekker Publication, 2004. |
 |
Tissue Engineering: Multi-Scaled Representation of Tissue Architecture and Function. Kaazempur-Mofrad MR, Weinberg EJ, Borenstein JT, Vacanti JP. in Complex Systems Science in Biomedicine (Eds.: Deisboeck TS, Kresh JY), Springer, NY, 2006. |
 |
Microvascular Engineering: Design, Modeling, and Microfabrication. Borenstein JT, Weinberg EJ, Vacanti JP, Mofrad MRK. in Micro- and Nanoengineering of the Cell Microenvironment, Technologies and Applications (Eds.: Khademhosseini et al.). Engineering in Medicine & Biology. 2008 |
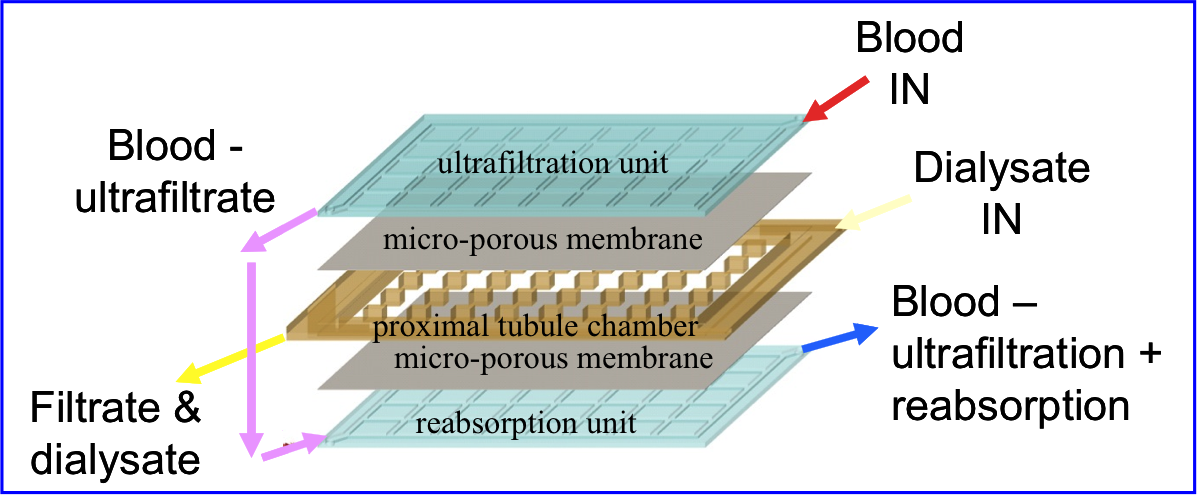 |
Technological Approaches to Renal Replacement Therapies. Reyes-Ortiz, Mofrad MRK, Weinberg EJ, Vacanti JP, Borenstein JT. in Micro- and Nanoengineering of the Cell Microenvironment, Technologies and Applications (Eds.: Khademhosseini et al.). Engineering in Medicine & Biology. 2008 |
 |
Computational modeling of aortic heart valve mechanics across multiple scales. Croft LR and Mofrad MRK. in Computational Cardiovascular Mechanics (Eds.: Guccione et al.). Springer, 2009 |
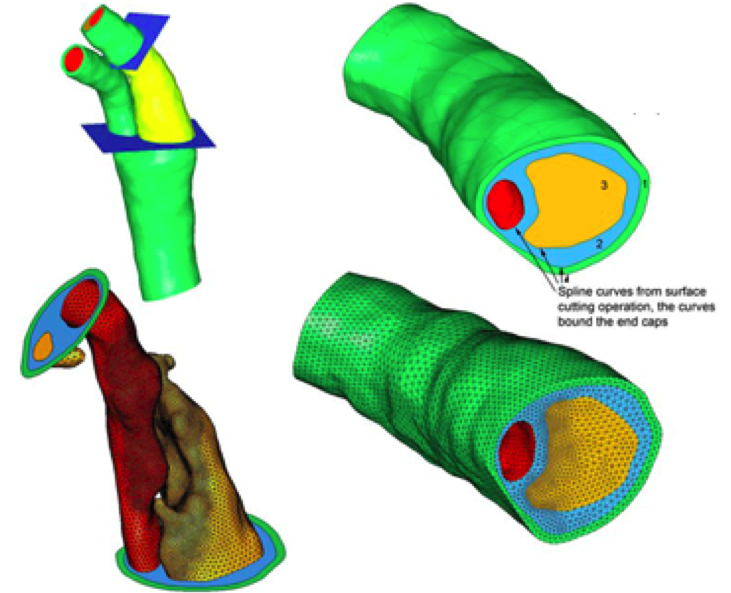 |
Computational models of vascular mechanics. Leach JR, Mofrad MRK, Saloner D. in Computational Modeling in Biomechanics (Eds.: De et al.). Springer, 2010 |
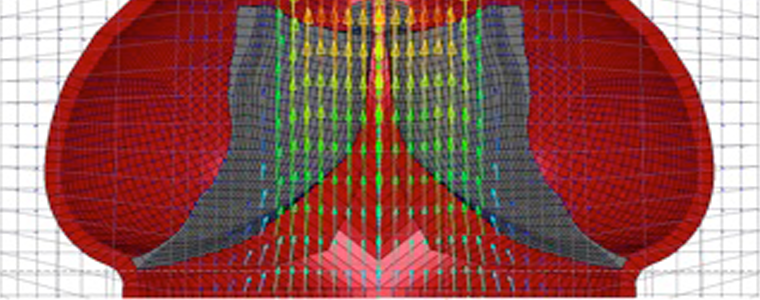 |
Computational modeling of aortic heart valves. Croft LR and Mofrad MRK. in Computational Modeling in Biomechanics (Eds.: De et al.). Springer, 2010 |
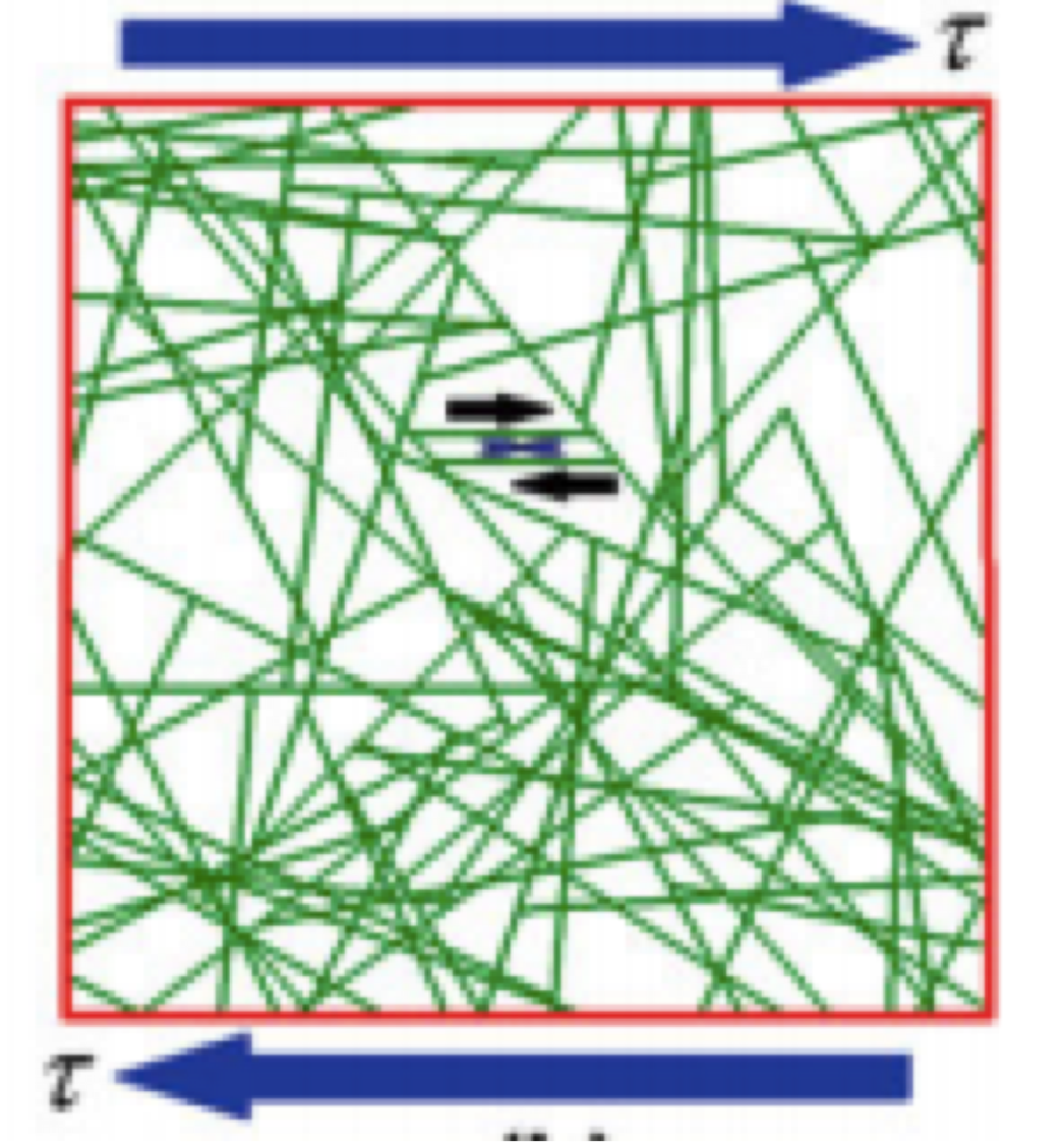 |
Cytoskeletal Mechanics and Cellular Mechanotransduction: A Molecular Perspective. Hatami H and Mofrad MRK. in Cellular and Biomolecular Mechanics and Mechanobiology (Ed.: Gefen A.). Springer, 2011 |
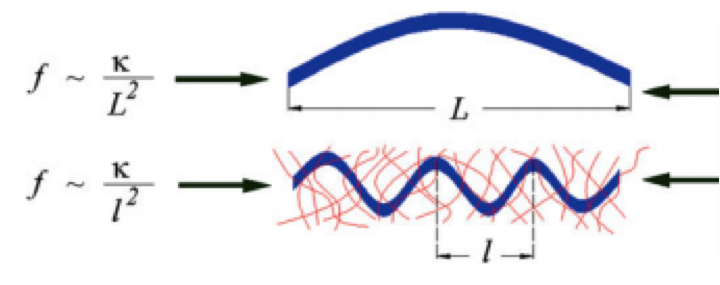 |
Cytoskeletal Mechanics and Rheology. Hatami H and Mofrad MRK. in Advances in Cell Mechanics (Eds.: Li S and Bohua S). Springer, 2011. |
 |
Mechanobiological Approaches for the Control of Cell Motility. Yoon SH, Mofrad MRK. in Microfluidic Cell Culture Systems (Eds.: Bettinger et al.), Elsevier, 2012. |
 |
Rheology and Mechanics of the Cell Cytoskeleton: A Complex Biopolymer Network Structure. Hatami H and Mofrad MRK. Complex Fluids in Biological Systems (Ed.: Spagnolie SE) Springer Biological and Medical Physics / Biological Engineering, In press. |


